1PML
 
 | |
1PMM
 
 | | Crystal structure of Escherichia coli GadB (low pH) | | 分子名称: | ACETIC ACID, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | 登録日 | 2003-06-11 | | 公開日 | 2004-02-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
1PMN
 
 | | Crystal structure of JNK3 in complex with an imidazole-pyrimidine inhibitor | | 分子名称: | CYCLOPROPYL-{4-[5-(3,4-DICHLOROPHENYL)-2-[(1-METHYL)-PIPERIDIN]-4-YL-3-PROPYL-3H-IMIDAZOL-4-YL]-PYRIMIDIN-2-YL}AMINE, Mitogen-activated protein kinase 10 | | 著者 | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | 登録日 | 2003-06-11 | | 公開日 | 2003-09-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
1PMO
 
 | | Crystal structure of Escherichia coli GadB (neutral pH) | | 分子名称: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate decarboxylase beta | | 著者 | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | 登録日 | 2003-06-11 | | 公開日 | 2004-02-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
1PMP
 
 | |
1PMR
 
 | | LIPOYL DOMAIN FROM THE DIHYDROLIPOYL SUCCINYLTRANSFERASE COMPONENT OF THE 2-OXOGLUTARATE DEHYDROGENASE MULTIENZYME COMPLEX OF ESCHERICHIA COLI, NMR, 25 STRUCTURES | | 分子名称: | DIHYDROLIPOYL SUCCINYLTRANSFERASE | | 著者 | Ricaud, P.M, Howard, M.J, Roberts, E.L, Broadhurst, R.W, Perham, R.N. | | 登録日 | 1997-07-24 | | 公開日 | 1998-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of the lipoyl domain from the dihydrolipoyl succinyltransferase component of the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli.
J.Mol.Biol., 264, 1996
|
|
1PMS
 
 | | PLECKSTRIN HOMOLOGY DOMAIN OF SON OF SEVENLESS 1 (SOS1) WITH GLYCINE-SERINE ADDED TO THE N-TERMINUS, NMR, 20 STRUCTURES | | 分子名称: | SOS 1 | | 著者 | Koshiba, S, Kigawa, T, Kim, J, Shirouzu, M, Bowtell, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1997-02-18 | | 公開日 | 1997-05-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the pleckstrin homology domain of mouse Son-of-sevenless 1 (mSos1).
J.Mol.Biol., 269, 1997
|
|
1PMT
 
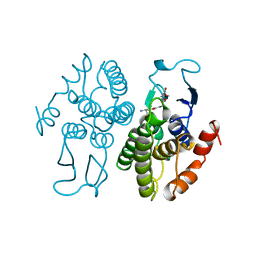 | | GLUTATHIONE TRANSFERASE FROM PROTEUS MIRABILIS | | 分子名称: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | 著者 | Rossjohn, J, Polekhina, G, Feil, S.C, Allocati, N, Masulli, M, Diilio, C, Parker, M.W. | | 登録日 | 1998-03-23 | | 公開日 | 1999-04-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications.
Structure, 6, 1998
|
|
1PMU
 
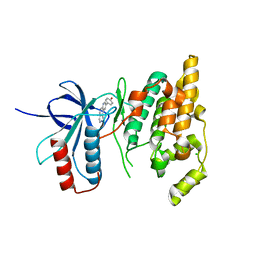 | | The crystal structure of JNK3 in complex with a phenantroline inhibitor | | 分子名称: | 9-(4-HYDROXYPHENYL)-2,7-PHENANTHROLINE, CHLORIDE ION, Mitogen-activated protein kinase 10 | | 著者 | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | 登録日 | 2003-06-11 | | 公開日 | 2003-09-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
1PMV
 
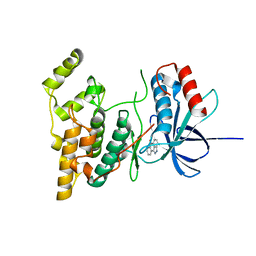 | | The structure of JNK3 in complex with a dihydroanthrapyrazole inhibitor | | 分子名称: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, Mitogen-activated protein kinase 10 | | 著者 | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | 登録日 | 2003-06-11 | | 公開日 | 2003-09-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
1PMX
 
 | |
1PMY
 
 | | REFINED CRYSTAL STRUCTURE OF PSEUDOAZURIN FROM METHYLOBACTERIUM EXTORQUENS AM1 AT 1.5 ANGSTROMS RESOLUTION | | 分子名称: | COPPER (II) ION, PSEUDOAZURIN | | 著者 | Inoue, T, Kai, Y, Harada, S, Kasai, N, Ohshiro, Y, Suzuki, S, Kohzuma, T, Tobari, J. | | 登録日 | 1994-01-28 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Refined crystal structure of pseudoazurin from Methylobacterium extorquens AM1 at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PN0
 
 | | Phenol hydroxylase from Trichosporon cutaneum | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHENOL, ... | | 著者 | Enroth, C. | | 登録日 | 2003-06-12 | | 公開日 | 2003-09-23 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High-resolution structure of phenol hydroxylase and correction of sequence errors.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PN2
 
 | | Crystal structure analysis of the selenomethionine labelled 2-enoyl-CoA hydratase 2 domain of Candida tropicalis multifunctional enzyme type 2 | | 分子名称: | 1,2-ETHANEDIOL, Peroxisomal hydratase-dehydrogenase-epimerase | | 著者 | Koski, M.K, Haapalainen, A.M, Hiltunen, J.K, Glumoff, T. | | 登録日 | 2003-06-12 | | 公開日 | 2004-04-13 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A Two-domain Structure of One Subunit Explains Unique Features of Eukaryotic Hydratase 2.
J.Biol.Chem., 279, 2004
|
|
1PN3
 
 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and the acceptor substrate DVV. | | 分子名称: | DESVANCOSAMINYL VANCOMYCIN, GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, ... | | 著者 | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | 登録日 | 2003-06-12 | | 公開日 | 2003-08-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1PN4
 
 | | Crystal structure of 2-enoyl-CoA hydratase 2 domain of Candida tropicalis multifunctional enzyme type 2 complexed with (3R)-hydroxydecanoyl-CoA. | | 分子名称: | 1,2-ETHANEDIOL, 3R-HYDROXYDECANOYL-COENZYME A, Peroxisomal hydratase-dehydrogenase-epimerase | | 著者 | Koski, M.K, Haapalainen, A.M, Hiltunen, J.K, Glumoff, T. | | 登録日 | 2003-06-12 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A Two-domain Structure of One Subunit Explains Unique Features of Eukaryotic Hydratase 2.
J.Biol.Chem., 279, 2004
|
|
1PN5
 
 | | NMR structure of the NALP1 Pyrin domain (PYD) | | 分子名称: | NACHT-, LRR- and PYD-containing protein 2 | | 著者 | Hiller, S, Kohl, A, Fiorito, F, Herrmann, T, Wider, G, Tschopp, J, Grutter, M.G, Wuthrich, K. | | 登録日 | 2003-06-12 | | 公開日 | 2003-10-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain
Structure, 11, 2003
|
|
1PN6
 
 | | Domain-wise fitting of the crystal structure of T.thermophilus EF-G into the low resolution map of the release complex.Puromycin.EFG.GDPNP of E.coli 70S ribosome. | | 分子名称: | Elongation factor G | | 著者 | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | 登録日 | 2003-06-12 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (10.8 Å) | | 主引用文献 | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1PN7
 
 | | Coordinates of S12, L11 proteins and P-tRNA, from the 70S X-ray structure aligned to the 70S Cryo-EM map of E.coli ribosome | | 分子名称: | 30S ribosomal protein S12, 50S ribosomal protein L11, P-tRNA | | 著者 | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | 登録日 | 2003-06-12 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (10.8 Å) | | 主引用文献 | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1PN8
 
 | | Coordinates of S12, L11 proteins and E-site tRNA from 70S crystal structure separately fitted into the Cryo-EM map of E.coli 70S.EF-G.GDPNP complex. The atomic coordinates originally from the E-site tRNA were fitted in the position of the hybrid P/E-site tRNA. | | 分子名称: | 30S ribosomal protein S12, 50S ribosomal protein L11, E-tRNA | | 著者 | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | 登録日 | 2003-06-12 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (10.8 Å) | | 主引用文献 | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1PN9
 
 | | Crystal structure of an insect delta-class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae | | 分子名称: | Glutathione S-transferase 1-6, S-HEXYLGLUTATHIONE | | 著者 | Chen, L, Hall, P.R, Zhou, X.E, Ranson, H, Hemingway, J, Meehan, E.J. | | 登録日 | 2003-06-12 | | 公開日 | 2003-12-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of an insect delta-class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PNB
 
 | | STRUCTURE OF NAPIN BNIB, NMR, 10 STRUCTURES | | 分子名称: | NAPIN BNIB | | 著者 | Rico, M, Bruix, M, Gonzalez, C, Monsalve, R, Rodriguez, R. | | 登録日 | 1996-09-17 | | 公開日 | 1997-09-17 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein.
Biochemistry, 35, 1996
|
|
1PNC
 
 | |
1PND
 
 | |
1PNE
 
 | | CRYSTALLIZATION AND STRUCTURE DETERMINATION OF BOVINE PROFILIN AT 2.0 ANGSTROMS RESOLUTION | | 分子名称: | PROFILIN | | 著者 | Cedergren-Zeppezauer, E.S, Goonesekere, N.C.W, Rozycki, M.D, Myslik, J.C, Dauter, Z, Lindberg, U, Schutt, C.E. | | 登録日 | 1995-05-05 | | 公開日 | 1995-07-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystallization and structure determination of bovine profilin at 2.0 A resolution.
J.Mol.Biol., 240, 1994
|
|
