6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
7NRZ
 
 | | Crystal structure of malate dehydrogenase from Trypanosoma cruzi | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sonani, R.R, Kurpiewska, K, Lewinski, K, Dubin, G. | | Deposit date: | 2021-03-04 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct sequence and structural feature of trypanosoma malate dehydrogenase.
Biochem.Biophys.Res.Commun., 557, 2021
|
|
4UUL
 
 | |
7F8D
 
 | | Malate Dehydrogenase from Geobacillus stearothermophilus (gs-MDH) G218Y mutant | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | Deposit date: | 2021-07-02 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Increasing loop flexibility affords low-temperature adaptation of a moderate thermophilic malate dehydrogenase from Geobacillus stearothermophilus.
Protein Eng.Des.Sel., 34, 2021
|
|
7QOZ
 
 | |
4UUM
 
 | |
4UUN
 
 | |
6TXR
 
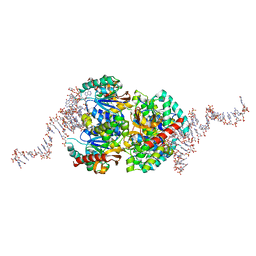 | | Structural insights into cubane-modified aptamer recognition of a malaria biomarker | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Cheung, Y, Roethlisberger, P, Mechaly, A, Weber, P, Wong, A, Lo, Y, Haouz, A, Savage, P, Hollenstein, M, Tanner, J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5A1T
 
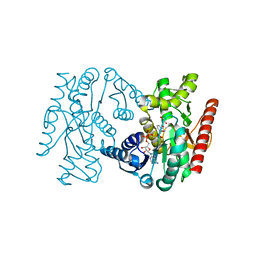 | | Trichomonas vaginalis lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-LACTATE DEHYDROGENASE, OXAMIC ACID | | Authors: | Steindel, P.A, Chen, E.H, Theobald, D.L. | | Deposit date: | 2015-05-05 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Gradual Neofunctionalization in the Convergent Evolution of Trichomonad Lactate and Malate Dehydrogenases.
Protein Sci., 25, 2016
|
|
7AOB
 
 | | Crystal structure of Thermaerobacter marianensis malate dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bertrand, Q, Lasalle, L, Girard, E, Madern, D. | | Deposit date: | 2020-10-14 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of Thermaerobacter marianensis malate dehydrogenase
To Be Published
|
|
6CT6
 
 | | Crystal structure of lactate dehydrogenase from Eimeria maxima with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Lactate dehydrogenase, OXAMIC ACID, ... | | Authors: | Wirth, J.D, Xu, C, Theobald, D.L. | | Deposit date: | 2018-03-22 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | The Mechanistic, Structural, and Evolutionary Origin of Lactate Dehydrogenase Substrate Specificity in Apicomplexa
To Be Published
|
|
7X1L
 
 | | Malate dehydrogenase from Geobacillus stearothermophilus (gs-MDH) delta E311 mutant complexed with Nicotinamide Adenine Dinucleotide (NAD+) | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | Deposit date: | 2022-02-24 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Reducing substrate inhibition of malate dehydrogenase from Geobacillus stearothermophilus by C-terminal truncation.
Protein Eng.Des.Sel., 35, 2022
|
|
6UM4
 
 | |
7W8A
 
 | | Babesia orientalis lactate dehydrogenase, BoLDH apo | | Descriptor: | lactate dehydrogenase | | Authors: | Yu, L. | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | The Structural Basis of Babesia orientalis Lactate Dehydrogenase.
Front Cell Infect Microbiol, 11, 2021
|
|
3FI9
 
 | | Crystal structure of malate dehydrogenase from Porphyromonas gingivalis | | Descriptor: | Malate dehydrogenase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-11 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of malate dehydrogenase from Porphyromonas gingivalis
To be Published
|
|
9JLO
 
 | | Crystal structure L-lactate dehydrogenase from Lactobacillus reuteri in its apoform | | Descriptor: | 1,2-ETHANEDIOL, L-lactate dehydrogenase | | Authors: | Matyuta, I.O, Minyaev, M.E, Shirokova, A.A, Pometun, A.A, Tishkov, V.I, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-09-19 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Novel extremely stable and active lactate dehydrogenase from l reuteri: structure-function studies and promising usage as an agent against pathogenic biofilms
To Be Published
|
|
7XQN
 
 | |
8AB3
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in complex with oxamate, NADH and FBP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,6-di-O-phosphono-beta-D-fructofuranose, L-lactate dehydrogenase, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8AB2
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in its apo form. | | Descriptor: | 1,2-ETHANEDIOL, L-lactate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
6R8G
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with 4-(3,4-difluorophenyl)thiazol-2-amine | | Descriptor: | 4-[3,4-bis(fluoranyl)phenyl]-1,3-thiazol-2-amine, Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fragment-Based Approach Identifies an Allosteric Pocket that Impacts Malate Dehydrogenase Activity
Commun Biol, 2021
|
|
6QSS
 
 | | Crystal Structure of Ignicoccus islandicus malate dehydrogenase co-crystallized with 10 mM Tb-Xo4 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Roche, J, Girard, E, Madern, D. | | Deposit date: | 2019-02-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | SOLUTION SCATTERING (1.892 Å), X-RAY DIFFRACTION | | Cite: | The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization.
J.Struct.Biol., 208, 2019
|
|
3OM9
 
 | |
5HTO
 
 | |
5HS4
 
 | | Plasmdoium Vivax Lactate dehydrogenase | | Descriptor: | L-lactate dehydrogenase | | Authors: | Choi, S.J, Ban, C. | | Deposit date: | 2016-01-25 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Crystal structure of a DNA aptamer bound to PvLDH elucidates novel single-stranded DNA structural elements for folding and recognition
Sci Rep, 6, 2016
|
|
5HRU
 
 | |
