8Q6W
 
 | |
8PKO
 
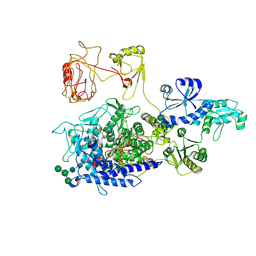 | | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Hitchman, C.J, Lia, A, Bayo, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer).
To Be Published
|
|
8V38
 
 | |
8T6K
 
 | |
8T6Q
 
 | |
8SYG
 
 | |
8T18
 
 | |
8T15
 
 | |
8T17
 
 | |
8JF7
 
 | |
8SFX
 
 | | High Affinity nanobodies against GFP | | Descriptor: | D-MALATE, GLYCEROL, Green fluorescent protein, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8SFV
 
 | | High affinity nanobodies to GFP | | Descriptor: | GLYCEROL, Green fluorescent protein, LaG19, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High affinity nanobodies to GFP
To Be Published
|
|
8SG3
 
 | |
8SFS
 
 | | High Affinity nanobodies against GFP | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonnano, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8SFZ
 
 | | High Affinity nanobodies against GFP | | Descriptor: | Green fluorescent protein, LaG35, POTASSIUM ION, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8SLC
 
 | | High Affinity nanobodies against GFP | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8J6O
 
 | | transport T2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Green fluorescent protein (Fragment),SID1 transmembrane family member 2, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J7H
 
 | | ion channel | | Descriptor: | ILE-ALA-ALA-ILE-HIS-ASN-ALA-ARG-ARG-LYS-LYS-ARG-GLU-ALA-ALA-ALA-ALA-HIS-LYS-ALA, ion channel | | Authors: | Chen, H.W, Jiang, D.H. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | ion channel
To Be Published
|
|
8TC3
 
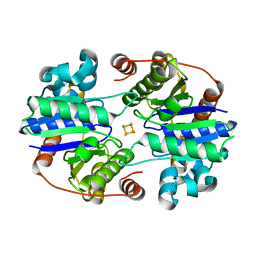 | |
8W2L
 
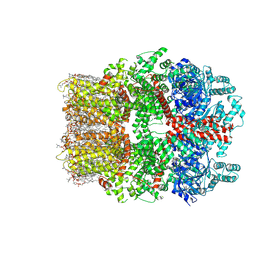 | | TRPM7 structure in complex with anticancer agent CCT128930 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, ... | | Authors: | Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2024-02-20 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural basis of selective TRPM7 inhibition by the anticancer agent CCT128930.
Cell Rep, 43, 2024
|
|
8XC6
 
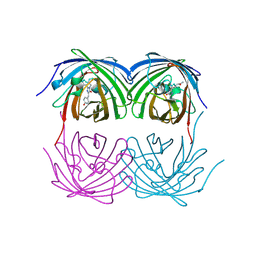 | |
8UBG
 
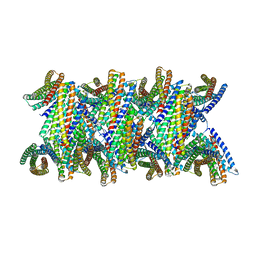 | | DpHF19 filament | | Descriptor: | DpHF19,Green fluorescent protein (Fragment) | | Authors: | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
8YDO
 
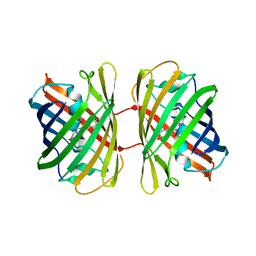 | | Crystal structure of dKeima570 | | Descriptor: | Large stokes shift fluorescent protein | | Authors: | Nam, K.H. | | Deposit date: | 2024-02-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dKeima570
To Be Published
|
|
8BBJ
 
 | |
8BAN
 
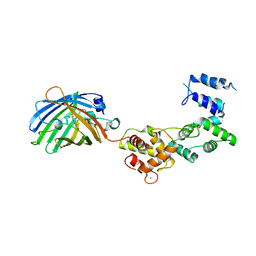 | | Secretagogin (mouse) in complex with its target peptide from SNAP-25 | | Descriptor: | CALCIUM ION, Green fluorescent protein,Synaptosomal-associated protein 25, Secretagogin | | Authors: | Schnell, R, Szodorai, E. | | Deposit date: | 2022-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A hydrophobic groove in secretagogin allows for alternate interactions with SNAP-25 and syntaxin-4 in endocrine tissues.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
