7DKA
 
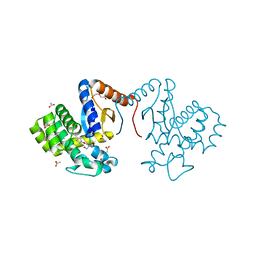 | | Crystal structure of DsbA-like protein DR2335 from Deinococcus radiodurans R1, C24S mutant protein | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, M.-K, Zhang, J, Zhao, L. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of DsbA-like protein DR2335 from Deinococcus radiodurans R1, native protein
To Be Published
|
|
5XWH
 
 | |
7PQ8
 
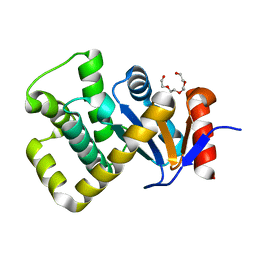 | | Crystal structure of Campylobacter jejuni DsbA1 | | Descriptor: | TETRAETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Orlikowska, M, Bocian-Ostrzycka, K.M, Banas, A.M, Jagusztyn-Krynicka, E.K. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
7PQ7
 
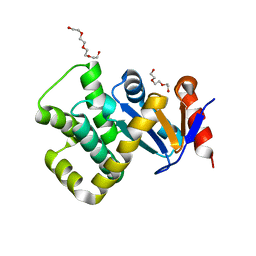 | | Crystal structure of Campylobacter jejuni DsbA1 | | Descriptor: | TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Wilk, P, Orlikowska, M, Banas, A.M, Bocian-Ostrzycka, K.M, Jagusztyn-Krynicka, E.K. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
3L9U
 
 | |
5CNW
 
 | |
5CO3
 
 | |
5CP1
 
 | |
5COH
 
 | |
5E59
 
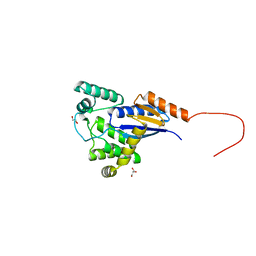 | |
4ZL9
 
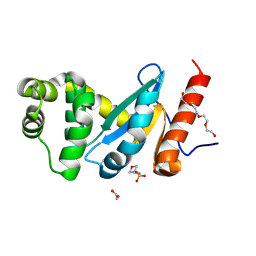 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal III | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
6WHD
 
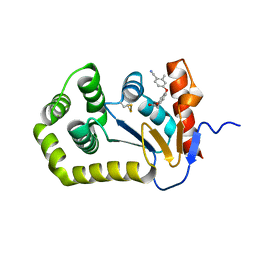 | | Crystal structure of E.coli DsbA in complex with diaryl ether analogue 2 | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, [4-(4-cyano-3-methylphenoxy)phenyl]acetic acid | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiLX).
J.Med.Chem., 63, 2020
|
|
2REM
 
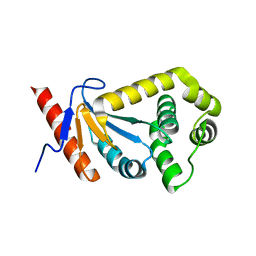 | |
6DXN
 
 | | 1.95 Angstrom Resolution Crystal Structure of DsbA Disulfide Interchange Protein from Klebsiella pneumoniae. | | Descriptor: | TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
5TLQ
 
 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
1R4W
 
 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
9EGR
 
 | | Crystal structure of oxidised E.coli DsbA in complex with allene | | Descriptor: | Thiol:disulfide interchange protein DsbA, methyl 4-{3-[(5-methyl-1,2-oxazole-3-carbonyl)amino]phenyl}butanoate | | Authors: | Balaji, G.R, Tasdan, Y, Ilyichova, O.V, Akhtar, N, Scanlon, M.J. | | Deposit date: | 2024-11-21 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of an Allene Warhead That Selectively Targets a Histidine Residue in the Escherichia coli Oxidoreductase Enzyme DsbA.
Acs Med.Chem.Lett., 16, 2025
|
|
4TKY
 
 | |
4DVC
 
 | | Structural and functional studies of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera toxin production | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Walden, P.M, Martin, J.L. | | Deposit date: | 2012-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A resolution crystal structure of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera-toxin production
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5VYO
 
 | |
5HFI
 
 | |
5HFG
 
 | | Cytosolic disulfide reductase DsbM from Pseudomonas aeruginosa | | Descriptor: | Uncharacterized protein, cytosolic disulfide reductase DsbM | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2016-01-07 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Crystal structures of the disulfide reductase DsbM from Pseudomonas aeruginosa
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7DK9
 
 | |
6BR4
 
 | | Crystal structure of Escherichia coli DsbA in complex with {N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
1FVK
 
 | |
