6GTT
 
 | | Human STK10 bound to BIRB-796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Serine/threonine-protein kinase 10 | | Authors: | Berger, B.T, Sorrell, F.J, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | STK10 bound to BIRB-796
To Be Published
|
|
9H4U
 
 | |
6CE6
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain soaked with 3,3'-(benzo[1,2-d:5,4-d']bis(thiazole)-2,6-diyl)dipropionic acid | | Descriptor: | 3,3'-(benzo[1,2-d:5,4-d']bis[1,3]thiazole-2,6-diyl)dipropanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
7Z89
 
 | | Sam68 | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Isoform 2 of KH domain-containing, ... | | Authors: | Nadal, M, Fuentes-Prior, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure and function analysis of Sam68 and hnRNP A1 synergy in the exclusion of exon 7 from SMN2 transcripts.
Protein Sci., 32, 2023
|
|
9BWS
 
 | |
4YR7
 
 | |
5LSZ
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
5J2Z
 
 | | PRV UL37 N-terminal half (R2 mutant) | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Heldwein, E.E, Pitts, J.D. | | Deposit date: | 2016-03-30 | | Release date: | 2017-10-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The pUL37 tegument protein guides alpha-herpesvirus retrograde axonal transport to promote neuroinvasion.
PLoS Pathog., 13, 2017
|
|
6MA4
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 3a | | Descriptor: | 5-{2-[(1R)-2-{(carboxymethyl)[(thiophen-2-yl)methyl]amino}-2-oxo-1-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}ethyl]phenoxy}pentanoic acid, Host Cell Factor 1 peptide, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
7V8J
 
 | |
6MBC
 
 | | Human Bfl-1 in complex with the designed peptide dF4 | | Descriptor: | Bcl-2-related protein A1, dF4 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
7KCP
 
 | |
6PL9
 
 | | Adduct formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimidaz ol-2-ylidene)(eta5-pentamethylcyclopentadienyl)rhodium(III) with HEWL | | Descriptor: | 2-(1-chloranyl-2,3,4,5,6-pentamethyl-1$l^{7}-rhodapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexan-1-yl)-1,3-dimethyl-benzimidazole, Lysozyme, SODIUM ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
6C3B
 
 | | O2-, PLP-Dependent L-Arginine Hydroxylase RohP Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
1JV6
 
 | | BACTERIORHODOPSIN D85S/F219L DOUBLE MUTANT AT 2.00 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
6WIX
 
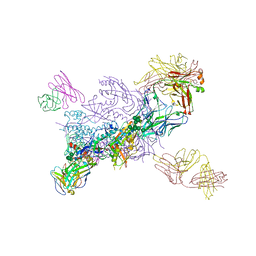 | | Crystal Structure of HIV-1 MI369 RnS-DS.SOSIP Prefusion Env Trimer in Complex with Human Antibodies 3H109L and 35O22 at 3.5 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Olia, A, Kwong, P.D. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Automated Design by Structure-Based Stabilization and Consensus Repair to Achieve Prefusion-Closed Envelope Trimers in a Wide Variety of HIV Strains.
Cell Rep, 33, 2020
|
|
6MES
 
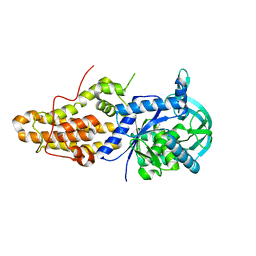 | |
6ZSD
 
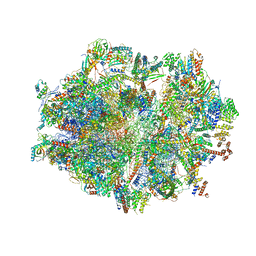 | | Human mitochondrial ribosome in complex with mRNA, P-site tRNA and E-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
8EXB
 
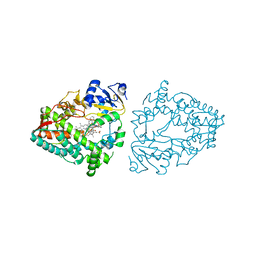 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-(pyridin-3-yl)propanamide}bis[2-(quinolin-2-yl-kappaN)-1-benzothiophen-3-yl-kappaC~3~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
6MA1
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 4a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2R)-2-{[(7-chloro-2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}-2-(2-methoxyphenyl)acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
5LP9
 
 | | FimA wt from S. flexneri | | Descriptor: | Major type 1 subunit fimbrin (Pilin) | | Authors: | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | Deposit date: | 2016-08-12 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.88626635 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
4Y5A
 
 | | Endothiapepsin in complex with fragment 206 | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-(trifluoromethyl)furan-2-yl]methanamine, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Huschmann, F.U, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
To Be Published
|
|
8BEO
 
 | | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, ... | | Authors: | Shaanan, B, Binshtein, E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP
Not Published
|
|
3NWP
 
 | |
8S6J
 
 | | NavMs in complex with riluzole | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 6-(trifluoromethoxy)-1,3-benzothiazol-2-amine, CHLORIDE ION, ... | | Authors: | Hollingworth, D, Sula, A, Mykhaylyk, V, Wallace, B.A. | | Deposit date: | 2024-02-27 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the rescue of hyperexcitable cells by the amyotrophic lateral sclerosis drug Riluzole.
Nat Commun, 15, 2024
|
|
