6FT8
 
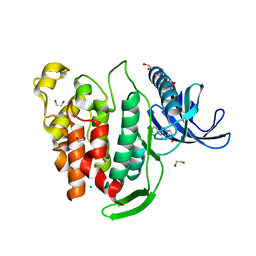 | | Crystal structure of CLK1 in complex with inhibitor 8g | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-hydroxyphenyl)-1~{H}-pyrrolo[3,4-g]indol-8-one, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
1KLJ
 
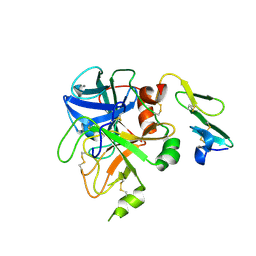 | | Crystal structure of uninhibited factor VIIa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, factor VIIa | | Authors: | Sichler, K, Banner, D, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions.
J.Mol.Biol., 322, 2002
|
|
6SNU
 
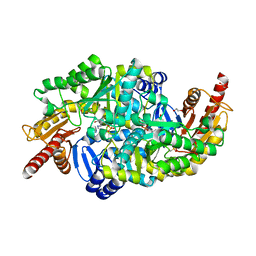 | | Crystal structure of the W60C mutant of the (S)-selective transaminase from Chromobacterium violaceum | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase family protein, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ruggieri, F, Gustafsson, C, Kimbung, R.Y, Walse, B, Logan, D.T, Berglund, P. | | Deposit date: | 2019-08-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Combined with Molecular Dynamics Reveal Altered Flow of Water in the Active Site of W60C Chromobacterium violaceum omega-transaminase
Not Published
|
|
6CQ0
 
 | | TBK1 in Complex with Dimethyl Amino Analog of Amlexanox | | Descriptor: | 2-amino-7-[3-(dimethylamino)propyl]-5-oxo-5H-[1]benzopyrano[2,3-b]pyridine-3-carboxylic acid, Serine/threonine-protein kinase TBK1 | | Authors: | Beyett, T.S, Tesmer, J.J.G. | | Deposit date: | 2018-03-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Design, synthesis, and biological activity of substituted 2-amino-5-oxo-5H-chromeno[2,3-b]pyridine-3-carboxylic acid derivatives as inhibitors of the inflammatory kinases TBK1 and IKK epsilon for the treatment of obesity.
Bioorg. Med. Chem., 26, 2018
|
|
5YN4
 
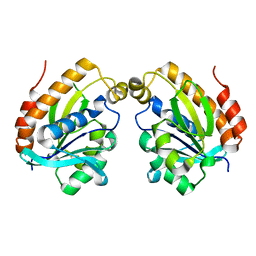 | |
9CCZ
 
 | | Crystal Structure of the K. pneumoniae LpxH / JH-LPH-92 Complex | | Descriptor: | 1-(5-{4-[6-chloro-4-(trifluoromethyl)pyridin-2-yl]piperazine-1-sulfonyl}-2,3-dihydro-1H-indol-1-yl)ethan-1-one, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Cochrane, C.S, Zhou, P. | | Deposit date: | 2024-06-23 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and Evaluation of Pyridinyl Sulfonyl Piperazine LpxH Inhibitors with Potent Antibiotic Activity Against Enterobacterales.
Jacs Au, 4, 2024
|
|
6CIW
 
 | |
6FQN
 
 | | Carbamylated T. californica acetylcholineterase bound to uncharged hybrid reactivator 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[4-[(7-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]butyl]-2-[(oxidanylamino)methyl]pyridin-3-ol, ... | | Authors: | De la Mora, E, Santoni, G, de Souza, J, Sussman, J, Silman, I, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.30002666 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
6STD
 
 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 3 | | Descriptor: | 2,2-DICHLORO-1-METHANESULFINYL-3-METHYL-CYCLOPROPANECARBOXYLIC ACID [1-(4-BROMO-PHENYL)-ETHYL]-AMIDE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-11 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
6SYR
 
 | | The wild type glucuronoyl esterase OtCE15A from Opitutus terrae in complex with D-glucuronate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Mazurkewich, S, Navarro Poulsen, J.C, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2019-10-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and biochemical studies of the glucuronoyl esteraseOtCE15A illuminate its interaction with lignocellulosic components.
J.Biol.Chem., 294, 2019
|
|
9CCX
 
 | | Crystal Structure of the Klebsiella pneumoniae LpxH/JH-LPH-86 complex | | Descriptor: | 1-(5-{4-[4-(trifluoromethyl)pyridin-2-yl]piperazine-1-sulfonyl}-2,3-dihydro-1H-indol-1-yl)ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cochrane, C.S, Zhou, P. | | Deposit date: | 2024-06-23 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Evaluation of Pyridinyl Sulfonyl Piperazine LpxH Inhibitors with Potent Antibiotic Activity Against Enterobacterales.
Jacs Au, 4, 2024
|
|
6LMR
 
 | | Solution structure of cold shock domain and ssDNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*CP*T)-3'), Y-box-binding protein 1 | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA binding to human YB-1 cold shock domain regulated by phosphorylation.
Nucleic Acids Res., 48, 2020
|
|
6FSY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Conway, S.J, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6SZ0
 
 | | The glucuronoyl esterase OtCE15A H408A variant from Opitutus terrae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Mazurkewich, S, Navarro Poulsen, J.C, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2019-10-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and biochemical studies of the glucuronoyl esteraseOtCE15A illuminate its interaction with lignocellulosic components.
J.Biol.Chem., 294, 2019
|
|
6FT7
 
 | | Crystal structure of CLK3 in complex with compound 8a | | Descriptor: | 1,2-ETHANEDIOL, 3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6FTM
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-048 | | Descriptor: | 1,2-ETHANEDIOL, 3-{5-[(4aR,8aS)-3-cycloheptyl-4-oxo-3,4,4a,5,8,8a-hexahydrophthalazin-1-yl]-2-methoxyphenyl}prop-2-ynamide, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-02-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-048
To be published
|
|
8W87
 
 | | Cryo-EM structure of the METH-TAAR1 complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
4Y4T
 
 | | Endothiapepsin in complex with fragment 114 | | Descriptor: | 1,2-ETHANEDIOL, 3-amino-5-(pyrrolidin-1-yl)-1H-pyrazole-4-carbonitrile, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
8W8A
 
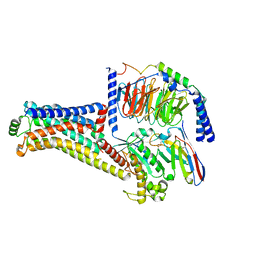 | | Cryo-EM structure of the RO5256390-TAAR1 complex | | Descriptor: | (4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-imine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8GD2
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-(2-thienylmethyl)methanamine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-[(thiophen-2-yl)methyl]methanamine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
3K50
 
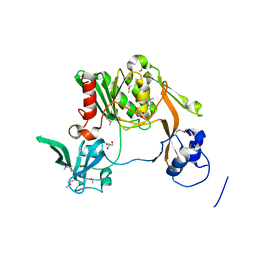 | |
6T5P
 
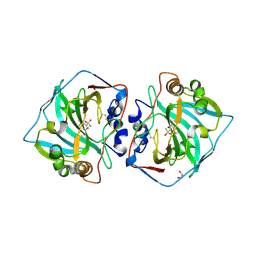 | | Human Carbonic anhydrase XII bound by 3,5-Di-tert-butylbenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 3,5-di~{tert}-butylbenzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-10-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Isoform-Selective Enzyme Inhibitors by Exploring Pocket Size According to the Lock-and-Key Principle.
Biophys.J., 119, 2020
|
|
6CSB
 
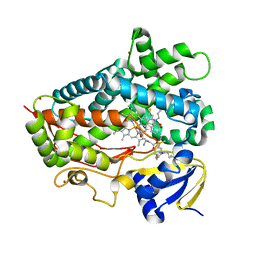 | | V308E mutant of cytochrome P450 2D6 complexed with thioridazine | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, ACETATE ION, Cytochrome P450 2D6, ... | | Authors: | Yang, Y.T, Fujita, K, Wang, P.F, Im, S.C, Pearl, N.M, Meagher, J, Stuckey, J, Waskell, L. | | Deposit date: | 2018-03-20 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Characteristic conformational changes on the distal and proximal surfaces of cytochrome P450 2D6 in response to substrate binding
To Be Published
|
|
6Z3I
 
 | | Structure of recombinant beta-glucocerebrosidase in complex with bifunctional cyclophellitol aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal acid glucosylceramidase, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-05-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
8EGY
 
 | | Engineered holo tyrosine synthase (TmTyrS1) derived from T. maritima TrpB | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
