6CF3
 
 | | Ethylene forming enzyme Y306A variant in complex with manganese and 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 2024
|
|
7K81
 
 | | KIR3DL1*005 in complex with HLA-A*24:02 presenting the RYPLTFGW peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ARG-TYR-PRO-LEU-THR-PHE-GLY-TRP, Beta-2-microglobulin, ... | | Authors: | MacLachlan, B.J, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Role of the HLA Class I alpha 2 Helix in Determining Ligand Hierarchy for the Killer Cell Ig-like Receptor 3DL1.
J Immunol., 206, 2021
|
|
7JU0
 
 | |
1LF9
 
 | | CRYSTAL STRUCTURE OF BACTERIAL GLUCOAMYLASE COMPLEXED WITH ACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE, SULFATE ION | | Authors: | Aleshin, A.E, Feng, P.-H, Honzatko, R.B, Reilly, P.J. | | Deposit date: | 2002-04-10 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and evolution of prokaryotic glucoamylase
J.Mol.Biol., 327, 2003
|
|
6CHN
 
 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with methyl (R)-4-(3-(2-cyano-1-((5-methyl-7-oxo-4,7-dihydro-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)ethyl)phenoxy)piperidine-1-carboxylate | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosphopantetheine adenylyltransferase, SULFATE ION, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
6CCM
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-((3-bromobenzyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one | | Descriptor: | 2-{[(3-bromophenyl)methyl]amino}-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6CCS
 
 | |
6CD2
 
 | | Crystal structure of the PapC usher bound to the chaperone-adhesin PapD-PapG | | Descriptor: | Chaperone protein PapD, Outer membrane usher protein PapC, PapGII adhesin protein | | Authors: | Omattage, N.S, Deng, Z, Yuan, P, Hultgren, S.J. | | Deposit date: | 2018-02-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for usher activation and intramolecular subunit transfer in P pilus biogenesis in Escherichia coli.
Nat Microbiol, 3, 2018
|
|
7G8U
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z131833926 | | Descriptor: | (3S)-3-{[(4R)-4-methyl-3,4-dihydropyridin-1(2H)-yl]methyl}-3H-indole, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
1HJQ
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-07-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1HKI
 
 | | Crystal structure of human chitinase in complex with glucoallosamidin B | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOTRIOSIDASE, METHYL N-ACETYL ALLOSAMINE | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase
J.Biol.Chem., 278, 2003
|
|
6CGP
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with MAIP-032 | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-hydroxy-4-[(2-propylimidazo[1,2-a]pyridin-3-yl)amino]benzamide, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2018-02-20 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multicomponent Synthesis and Binding Mode of Imidazo[1,2- a]pyridine-Capped Selective HDAC6 Inhibitors.
Org. Lett., 20, 2018
|
|
1K01
 
 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, CHLORAMPHENICOL, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
6CIC
 
 | | Structure of the human nitric oxide synthase R354A/G357D mutant heme domain in complex with N-(1-(2-(Ethyl(methyl)amino)ethyl)-1,2,3,4-tetrahydroquino-lin-6-yl)thiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, N-(1-{2-[ethyl(methyl)amino]ethyl}-1,2,3,4-tetrahydroquinolin-6-yl)thiophene-2-carboximidamide, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structural Basis for Isoform Selective Nitric Oxide Synthase Inhibition by Thiophene-2-carboximidamides.
Biochemistry, 57, 2018
|
|
6CIF
 
 | | Structure of the human endothelial nitric oxide synthase heme domain in complex with N-(1-(Piperidin-4-yl)indolin-5-yl)thiophene-2-carboximidamide | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Isoform Selective Nitric Oxide Synthase Inhibition by Thiophene-2-carboximidamides.
Biochemistry, 57, 2018
|
|
6CIQ
 
 | |
1JZZ
 
 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, MAGNESIUM ION, ROXITHROMYCIN, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
6CXI
 
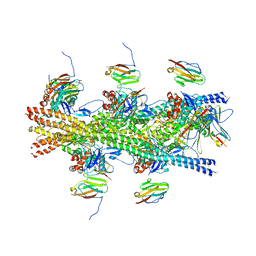 | |
1GED
 
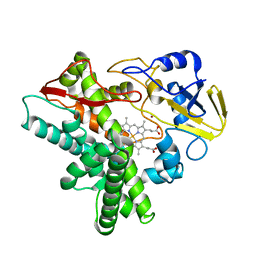 | | A positive charge route for the access of nadh to heme formed in the distal heme pocket of cytochrome p450nor | | Descriptor: | BROMIDE ION, CYTOCHROME P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kudo, T, Takaya, N, Park, S.-Y, Shiro, Y, Shoun, H. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A positively charged cluster formed in the heme-distal pocket of cytochrome P450nor is essential for interaction with NADH
J.Biol.Chem., 276, 2001
|
|
1GDK
 
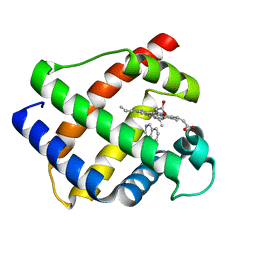 | |
1JYA
 
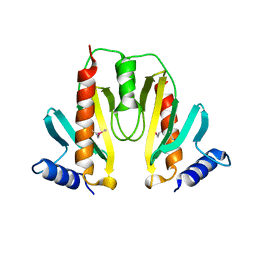 | | Crystal Structure of SycE | | Descriptor: | YOPE regulator | | Authors: | Ghosh, P, Birtalan, S. | | Deposit date: | 2001-09-11 | | Release date: | 2001-10-31 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the Yersinia type III secretory system chaperone SycE.
Nat.Struct.Biol., 8, 2001
|
|
1JZ4
 
 | |
1K0O
 
 | | Crystal structure of a soluble form of CLIC1. An intracellular chloride ion channel | | Descriptor: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1 | | Authors: | Harrop, S.J, DeMaere, M.Z, Fairlie, W.D, Reztsova, T, Valenzuela, S.M, Mazzanti, M, Tonini, R, Qiu, M.R, Jankova, L, Warton, K, Bauskin, A.R, Wu, W.M, Pankhurst, S, Campbell, T.J, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution.
J.Biol.Chem., 276, 2001
|
|
1GH0
 
 | | CRYSTAL STRUCTURE OF C-PHYCOCYANIN FROM SPIRULINA PLATENSIS | | Descriptor: | C-PHYCOCYANIN ALPHA SUBUNIT, C-PHYCOCYANIN BETA SUBUNIT, PHYCOCYANOBILIN | | Authors: | Liang, D.-C, Chang, W.-R, Wang, X.-Q. | | Deposit date: | 2000-10-29 | | Release date: | 2001-06-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of C-phycocyanin from Spirulina platensis at 2.2 A resolution: a novel monoclinic crystal form for phycobiliproteins in phycobilisomes.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1K2R
 
 | | Structure of rat brain nNOS heme domain complexed with NG-nitro-L-arginine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-OMEGA-NITRO-L-ARGININE, ... | | Authors: | Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2001-09-28 | | Release date: | 2003-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of rat brain nNOS heme domain
To be Published
|
|
