8DQI
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to ATP and acridone after 2- weeks of crystal growth | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
8DQH
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to ATP and acridone after 24 hours of crystal growth | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
8DQJ
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (AST) bound to ATP and acridone | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
1V7O
 
 | |
1USY
 
 | | ATP phosphoribosyl transferase (HisG:HisZ) complex from Thermotoga maritima | | Descriptor: | ATP PHOSPHORIBOSYLTRANSFERASE, ATP PHOSPHORIBOSYLTRANSFERASE REGULATORY SUBUNIT, HISTIDINE, ... | | Authors: | Vega, M.C, Fernandez, F.J, Murphy, G.E, Zou, P, Popov, A, Wilmanns, M. | | Deposit date: | 2003-12-01 | | Release date: | 2004-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Regulation of the Hetero-Octameric ATP Phosphoribosyl Transferase Complex from Thermotoga Maritima by a tRNA Synthetase-Like Subunit.
Mol.Microbiol., 55, 2005
|
|
5O5Z
 
 | | CRYSTAL STRUCTURE OF THERMOCOCCUS LITORALIS ADP-DEPENDENT GLUCOKINASE (GK) | | Descriptor: | 5'-O-[(R)-HYDROXY(THIOPHOSPHONOOXY)PHOSPHORYL]ADENOSINE, ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, ... | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5O5Y
 
 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5O5X
 
 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, SULFATE ION | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
7OTE
 
 | | Src Kinase Domain in complex with ponatinib | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, GLYCEROL, ... | | Authors: | Soriano-Maldonado, P, Cuesta-Hernandez, H.N, Plaza-Menacho, I. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric switch between the activation loop and a c-terminal palindromic phospho-motif controls c-Src function.
Nat Commun, 14, 2023
|
|
1DSY
 
 | | C2 DOMAIN FROM PROTEIN KINASE C (ALPHA) COMPLEXED WITH CA2+ AND PHOSPHATIDYLSERINE | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Verdaguer, N, Corbalan-Garcia, S, Ochoa, W.F, Fita, I, Gomez-Fernandez, J.C. | | Deposit date: | 2000-01-10 | | Release date: | 2000-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine.
EMBO J., 18, 1999
|
|
8TRD
 
 | | mGluR3 class 1 in the presence of the antagonist LY 341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, Metabotropic glutamate receptor 3 | | Authors: | Strauss, A, Levitz, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of positive allosteric modulation of metabotropic glutamate receptor activation and internalization.
Nat Commun, 15, 2024
|
|
4APY
 
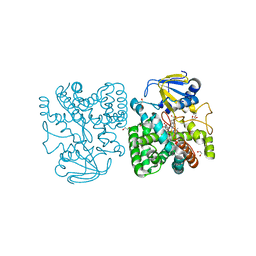 | | Ethylene glycol-bound form of P450 CYP125A3 from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Frank, D.J, Garcia Fernandez, E, Kells, P.M, Garcia Lopez, J.L, Podust, L.M, Ortiz de Montellano, P.R. | | Deposit date: | 2012-04-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
4CD6
 
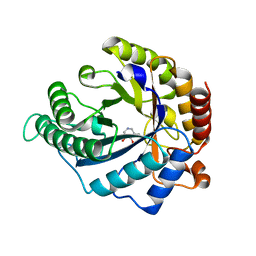 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManIFG | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD8
 
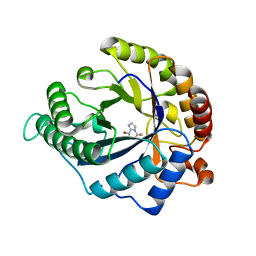 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManMIm | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD7
 
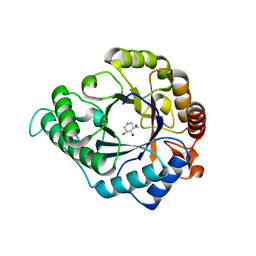 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManIFG and beta-1,4-mannobiose | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD5
 
 | | The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManMIm | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ENDO-1,4-BETA MANNANASE, PUTATIVE, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD4
 
 | | The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManIFG | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-1,4-BETA MANNANASE, PUTATIVE, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8RPG
 
 | | Crystal structure of an alcohol oxidase from Streptomyces hiroshimensis | | Descriptor: | Alcohol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martinez-Julvez, M, Ferreira, P, Cinca-Fernando, P. | | Deposit date: | 2024-01-15 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery, characterization, and synthetic potential of two novel bacterial aryl-alcohol oxidases.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
8PE1
 
 | | Crystal structure of Gel4 in complex with Nanobody 4 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 4, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8PE2
 
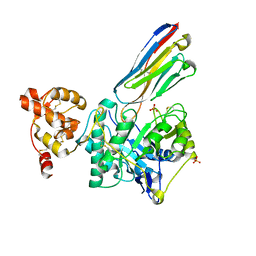 | | Crystal structure of Gel4 in complex with Nanobody 3 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 3, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
3G19
 
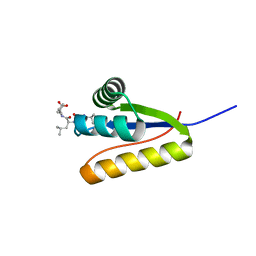 | | The structure of the Caulobacter crescentus clpS protease adaptor protein in complex with LLL tripeptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, LLL tripeptide | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G3P
 
 | |
3G1B
 
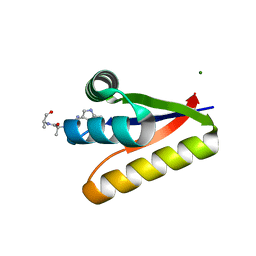 | | The structure of the M53A mutant of Caulobacter crescentus clpS protease adaptor protein in complex with WLFVQRDSKE peptide | | Descriptor: | 10-residue peptide, ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GQ1
 
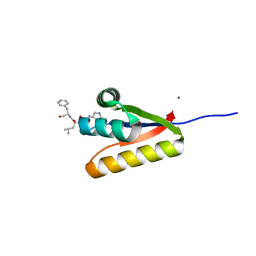 | | The structure of the caulobacter crescentus clpS protease adaptor protein in complex with a WLFVQRDSKE decapeptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, WLFVQRDSKE peptide | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GQ0
 
 | |
