7B2R
 
 | |
7B36
 
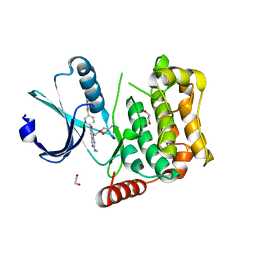 | | MST4 in complex with compound G-5555 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, CHLORIDE ION, ... | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.10681081 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
6J0Y
 
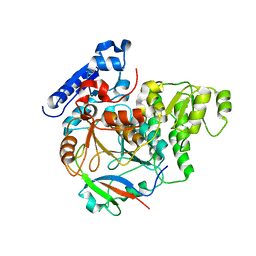 | | Crystal Structure of Yeast Rtt107 and Slx4 | | Descriptor: | Peptide from Structure-specific endonuclease subunit SLX4, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J1J
 
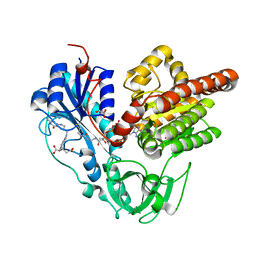 | | Crystal structure of HypX from Aquifex aeolicus, A392F-I419F variant in complex with Tetrahydrofolic acid | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, COENZYME A, GLYCEROL, ... | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of HypX responsible for CO biosynthesis in the maturation of NiFe-hydrogenase.
Commun Biol, 2, 2019
|
|
7M8S
 
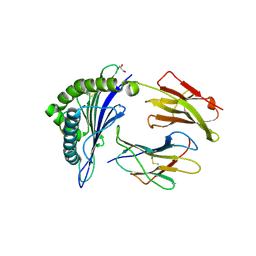 | | Crystal Structure of HLA-A*02:01 in complex with KLNDLCFTNV, an 10-mer epitope from SARS-CoV-2 Spike (S386-395) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | SARS-CoV-2 Spike-Derived Peptides Presented by HLA Molecules
Biophysica, 1, 2021
|
|
7ZL8
 
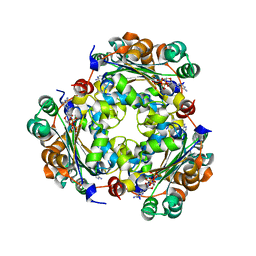 | | NME1 in complex with succinyl-CoA | | Descriptor: | Nucleoside diphosphate kinase A, SUCCINYL-COENZYME A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
7MGS
 
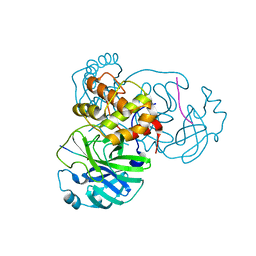 | |
7RMQ
 
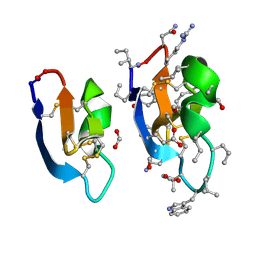 | | Crystal structure of cycloviolacin O2 | | Descriptor: | Cycloviolacin O2, D-[I11L]cycloviolacin O2, FORMIC ACID | | Authors: | Huang, Y.H, Du, Q. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
8SXQ
 
 | |
7RMS
 
 | | Crystal structure of [I11G]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
7ZTN
 
 | | Crystal structure of fungal CE16 acetyl xylan esterase | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Kosinas, C, Pentari, C, Zerva, A, Topakas, E. | | Deposit date: | 2022-05-10 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The role of CE16 exo-deacetylases in hemicellulolytic enzyme mixtures revealed by the biochemical and structural study of the novel TtCE16B esterase.
Carbohydr Polym, 327, 2024
|
|
6D43
 
 | |
7B7N
 
 | | Human herpesvirus-8 gH/gL in complex with EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Pederzoli, R, Guardado-Calvo, P, Rey, F.A, Backovic, M. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Human herpesvirus 8 molecular mimicry of ephrin ligands facilitates cell entry and triggers EphA2 signaling.
Plos Biol., 19, 2021
|
|
7M8U
 
 | | Crystal Structure of HLA-B*35:01 in complex with IPFAMQMAY, an 9-mer epitope from SARS-CoV-2 spike (S896-904) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen B, alpha chain, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SARS-CoV-2 Spike-Derived Peptides Presented by HLA Molecules
Biophysica, 1, 2021
|
|
7ZLW
 
 | | NME1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-04-15 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
6D4R
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 18 (VCC399134) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, hydroxy(3-{4-[(isoquinolin-5-yl)sulfonyl]piperazine-1-carbonyl}phenyl)oxoammonium | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
7RKS
 
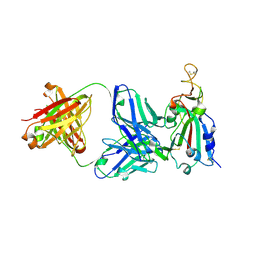 | | Structure of the SARS-CoV receptor binding domain in complex with the human neutralizing antibody Fab fragment, C118 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Antibody Fab Heavy Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
6IXR
 
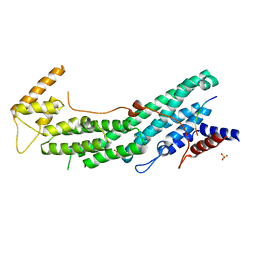 | | Structure of Myo2-GTD in complex with Inp2 | | Descriptor: | Inheritance of peroxisomes protein 2, Myosin-2, SULFATE ION | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
7RNK
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-71 | | Descriptor: | 3C-like proteinase, 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7B88
 
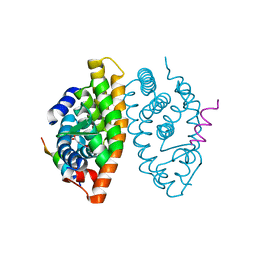 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with S99 inhibitor | | Descriptor: | 3-[5-[3,5-bis(chloranyl)phenyl]-4-phenyl-1,3-oxazol-2-yl]propanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Schierle, S, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Oxaprozin Analogues as Selective RXR Agonists with Superior Properties and Pharmacokinetics.
J.Med.Chem., 64, 2021
|
|
7M8T
 
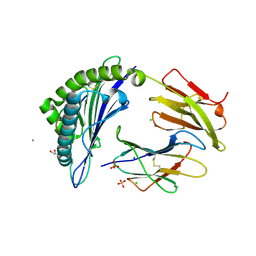 | | Crystal Structure of HLA-A*11:01 in complex with NSASFSTFK, an 9-mer epitope from SARS-CoV-2 spike (S370-378) | | Descriptor: | Beta-2-microglobulin, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | SARS-CoV-2 Spike-Derived Peptides Presented by HLA Molecules
Biophysica, 1, 2021
|
|
7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | Descriptor: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RME
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-52 | | Descriptor: | 3C-like proteinase, 6-{4-[4-chloro-3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7ZG3
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011228 | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-[(1~{S})-1,2,2-trimethylcyclopropyl]pyrrolo[1,2-c]pyrimidine-3-carboxamide | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-04-01 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011228
To Be Published
|
|
8A0S
 
 | | MTH1 in complex with TH013350 | | Descriptor: | 4-[(2-chlorophenyl)methylsulfanyl]-5~{H}-pyrimidin-2-one, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Liu, H, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-05-30 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | MTH1 in complex with TH013350
To Be Published
|
|
