8J7E
 
 | | Crystal structure of BRIL in complex with 1b3 Fab | | Descriptor: | Antibody 1b3 Fab Heavy chain, Antibody 1b3 Fab Light chain, Soluble cytochrome b562 | | Authors: | Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8J7W
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 2, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-bound conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
1FHE
 
 | | GLUTATHIONE TRANSFERASE (FH47) FROM FASCIOLA HEPATICA | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 1997-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization, structural determination and analysis of a novel parasite vaccine candidate: Fasciola hepatica glutathione S-transferase.
J.Mol.Biol., 273, 1997
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
8IAJ
 
 | | Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A) complex | | Descriptor: | N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, PYRIDOXAL-5'-PHOSPHATE, Protein ORM2, ... | | Authors: | Xie, T, Gong, X. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide.
Cell Rep, 43, 2024
|
|
8I9C
 
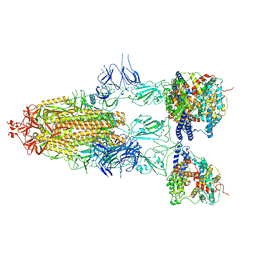 | | S-ECD (Omicron BF.7) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9D
 
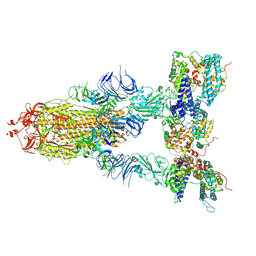 | | S-ECD (Omicron XBB.1) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
1QNO
 
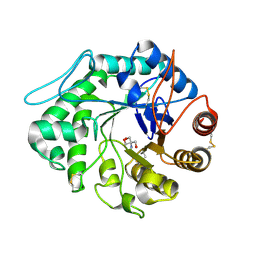 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
8IAM
 
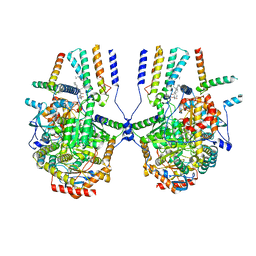 | | Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3D) complex | | Descriptor: | Chimera of Long chain base biosynthesis protein 1 and Serine palmitoyltransferase 1, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Xie, T, Gong, X. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide.
Cell Rep, 43, 2024
|
|
8I94
 
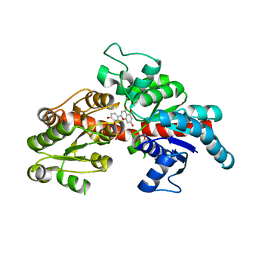 | | Structure of flavone 4'-O-glucoside 7-O-glucosyltransferase from Nemophila menziesii, complex with luteolin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, Glycosyltransferase, SULFATE ION | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Molecular basis of ligand recognition specificity of flavone glucosyltransferases in Nemophila menziesii.
Arch.Biochem.Biophys., 753, 2024
|
|
8I90
 
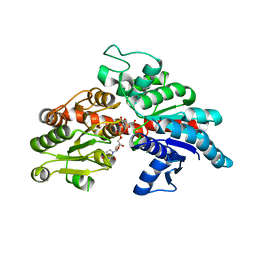 | |
1FFS
 
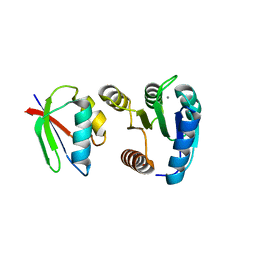 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY FROM CRYSTALS SOAKED IN ACETYL PHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1QV1
 
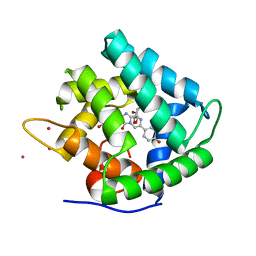 | | Atomic resolution structure of obelin from Obelia longissima | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Liu, Z.J, Vysotski, E.S, Deng, L, Lee, J, Rose, J, Wang, B.C. | | Deposit date: | 2003-08-26 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structure of obelin: soaking with calcium enhances electron density of the second oxygen atom substituted at the C2-position of coelenterazine.
Biochem.Biophys.Res.Commun., 311, 2003
|
|
1FKR
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKO
 
 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-10 | | Release date: | 2000-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
1FLG
 
 | | CRYSTAL STRUCTURE OF THE QUINOPROTEIN ETHANOL DEHYDROGENASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | CALCIUM ION, PROTEIN (QUINOPROTEIN ETHANOL DEHYDROGENASE), PYRROLOQUINOLINE QUINONE | | Authors: | Keitel, T, Diehl, A, Knaute, T, Stezowski, J.J, Hohne, W, Gorisch, H. | | Deposit date: | 2000-08-14 | | Release date: | 2000-08-30 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the quinoprotein ethanol dehydrogenase from Pseudomonas aeruginosa: basis of substrate specificity.
J.Mol.Biol., 297, 2000
|
|
1QHZ
 
 | |
1FN3
 
 | | CRYSTAL STRUCTURE OF NICKEL RECONSTITUTED HEMOGLOBIN-A CASE FOR PERMANENT, T-STATE HEMOGLOBIN | | Descriptor: | HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, PROTOPORPHYRIN IX CONTAINING NI(II) | | Authors: | Venkateshrao, S, Deepthi, S, Pattabhi, V, Manoharan, P.T. | | Deposit date: | 2000-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of Nickel Reconstituted Hemoglobin - A Case for Permanent, T-State Hemoglobin
CURR.SCI., 84, 2003
|
|
8IE4
 
 | |
1FOI
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 1400W(H4B-FREE) | | Descriptor: | ACETATE ION, CACODYLATE ION, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-08-28 | | Release date: | 2001-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic studies on endothelial nitric oxide synthase complexed with nitric oxide and mechanism-based inhibitors.
Biochemistry, 40, 2001
|
|
1FFE
 
 | |
1FQ6
 
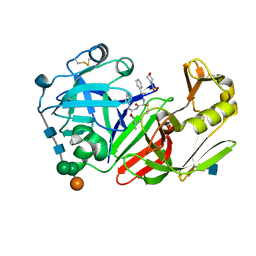 | | X-RAY STRUCTURE OF GLYCOL INHIBITOR PD-133,450 BOUND TO SACCHAROPEPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1S)-2-{[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]amino}-1-(ethylsulfanyl)-2-oxoethyl]-Nalpha-(morp holin-4-ylsulfonyl)-L-phenylalaninamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
1Q8U
 
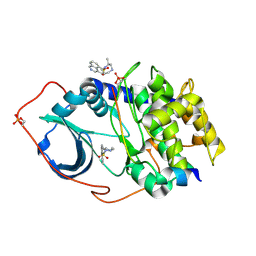 | | The Catalytic Subunit of cAMP-dependent Protein Kinase in Complex with Rho-kinase Inhibitor H-1152P | | Descriptor: | (S)-2-METHYL-1-[(4-METHYL-5-ISOQUINOLINE)SULFONYL]-HOMOPIPERAZINE, N-OCTANOYL-N-METHYLGLUCAMINE, cAMP-dependent protein kinase inhibitor, ... | | Authors: | Breitenlechner, C, Gassel, M, Hidaka, H, Kinzel, V, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: structural basis of selectivity.
Structure, 11, 2003
|
|
1QNP
 
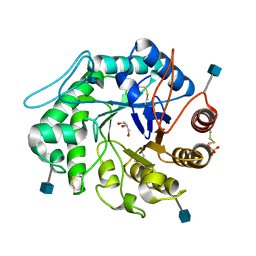 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, GLYCEROL, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1Q9G
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX
J.Mol.Biol., 336, 2004
|
|
