1NDH
 
 | |
1NK0
 
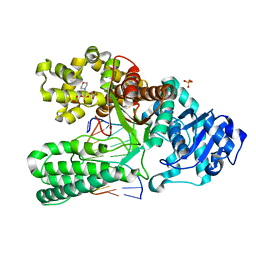 | | ADENINE-GUANINE MISMATCH AT THE POLYMERASE ACTIVE SITE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA POLYMERASE I, DNA PRIMER STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NKV
 
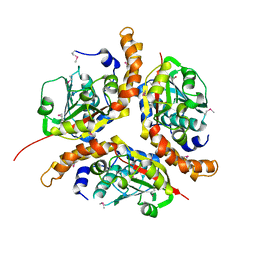 | | X-RAY STRUCTURE OF YJHP FROM E.COLI NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER13 | | Descriptor: | HYPOTHETICAL PROTEIN yjhP | | Authors: | Kuzin, A, Manor, P, Benach, J, Smith, P, Rost, B, Xiao, R, Montelione, G, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-03 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-RAY STRUCTURE OF YJHP FROM E.COLI NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER13
To be published
|
|
1NL2
 
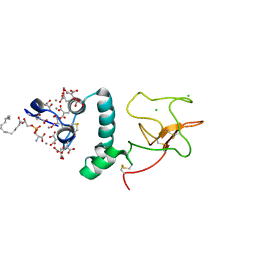 | | BOVINE PROTHROMBIN FRAGMENT 1 IN COMPLEX WITH CALCIUM AND LYSOPHOSPHOTIDYLSERINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, M, Huang, G, Furie, B, Seaton, B, Furie, B.C. | | Deposit date: | 2003-01-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of membrane binding by Gla domains of vitamin K-dependent proteins.
Nat.Struct.Biol., 10, 2003
|
|
4FMG
 
 | |
1NJW
 
 | |
1NDI
 
 | | Carnitine Acetyltransferase in complex with CoA | | Descriptor: | COENZYME A, Carnitine Acetyltransferase | | Authors: | Jogl, G, Tong, L. | | Deposit date: | 2002-12-09 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Carnitine Acetyltransferase and Implications for the Catalytic Mechanism and Fatty Acid Transport
Cell(Cambridge,Mass.), 112, 2003
|
|
1NK5
 
 | |
1NHU
 
 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
1NIT
 
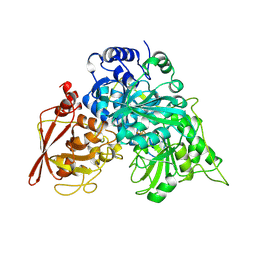 | | CRYSTAL STRUCTURE OF ACONITASE WITH TRANS-ACONITATE AND NITROCITRATE BOUND | | Descriptor: | ACONITASE, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Lauble, H, Kennedy, M.C, Beinert, H, Stout, C.D. | | Deposit date: | 1993-01-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of aconitase with trans-aconitate and nitrocitrate bound.
J.Mol.Biol., 237, 1994
|
|
1NIH
 
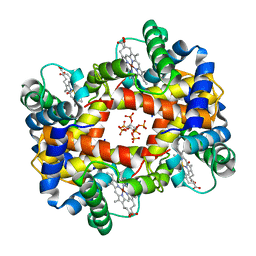 | |
1N7F
 
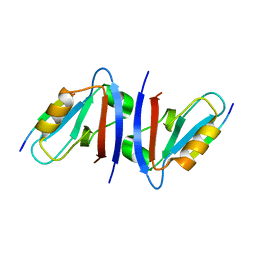 | | Crystal structure of the sixth PDZ domain of GRIP1 in complex with liprin C-terminal peptide | | Descriptor: | 8-mer peptide from interacting protein (liprin), AMPA receptor interacting protein GRIP | | Authors: | Im, Y.J, Park, S.H, Rho, S.H, Lee, J.H, Kang, G.B, Sheng, M, Kim, E, Eom, S.H. | | Deposit date: | 2002-11-14 | | Release date: | 2003-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization
J.BIOL.CHEM., 278, 2003
|
|
1N7V
 
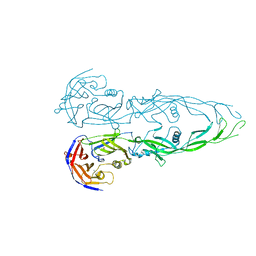 | | THE RECEPTOR-BINDING PROTEIN P2 OF BACTERIOPHAGE PRD1: CRYSTAL FORM III | | Descriptor: | ACETATE ION, Adsorption protein P2, CALCIUM ION | | Authors: | Xu, L, Benson, S.D, Butcher, S.J, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-11-18 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Receptor Binding Protein P2 of PRD1, a
Virus Targeting Antibiotic-Resistant Bacteria,
Has a Novel Fold Suggesting Multiple Functions.
Structure, 11, 2003
|
|
1NCA
 
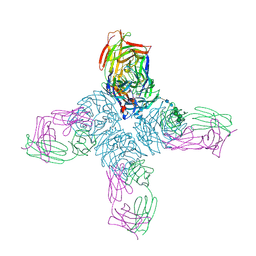 | | REFINED CRYSTAL STRUCTURE OF THE INFLUENZA VIRUS N9 NEURAMINIDASE-NC41 FAB COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex.
J.Mol.Biol., 227, 1992
|
|
1NG1
 
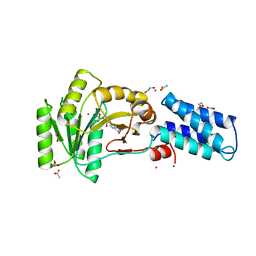 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CADMIUM ION, ... | | Authors: | Freymann, D.M, Stroud, R.M, Walter, P. | | Deposit date: | 1998-04-30 | | Release date: | 1999-07-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP.
Nat.Struct.Biol., 6, 1999
|
|
1E7I
 
 | |
1NAP
 
 | |
1NBZ
 
 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K97A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
1NCB
 
 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
1NCH
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NED
 
 | |
1NF5
 
 | | Crystal Structure of Lactose Synthase, Complex with Glucose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-lactalbumin, CALCIUM ION, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2002-12-13 | | Release date: | 2002-12-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lactose Synthase Reveals a Large Conformational Change in its Catalytic Component, the beta-1,4-galactosyltransferase
J.Mol.Biol., 310, 2001
|
|
1NFK
 
 | | STRUCTURE OF THE NUCLEAR FACTOR KAPPA-B (NF-KB) P50 HOMODIMER | | Descriptor: | DNA (5'-D(*TP*GP*GP*GP*AP*AP*TP*TP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B (NF-KB)) | | Authors: | Ghosh, G, Van Duyne, G, Ghosh, S, Sigler, P.B. | | Deposit date: | 1995-02-28 | | Release date: | 1996-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of NF-kappa B p50 homodimer bound to a kappa B site.
Nature, 373, 1995
|
|
1NK7
 
 | |
1NL0
 
 | | Crystal structure of human factor IX Gla domain in complex of an inhibitory antibody, 10C12 | | Descriptor: | CALCIUM ION, SULFATE ION, anti-factor IX antibody, ... | | Authors: | Huang, M, Furie, B.C, Furie, B. | | Deposit date: | 2003-01-06 | | Release date: | 2004-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Calcium-stabilized Human Factor IX Gla Domain Bound to a Conformation-specific Anti-factor IX Antibody.
J.Biol.Chem., 279, 2004
|
|
