3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | Descriptor: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | Authors: | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
1KR2
 
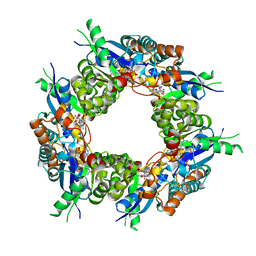 | | CRYSTAL STRUCTURE OF HUMAN NMN/NAMN ADENYLYL TRANSFERASE COMPLEXED WITH TIAZOFURIN ADENINE DINUCLEOTIDE (TAD) | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
6GGL
 
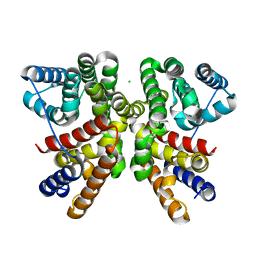 | | Crystal structure of CotB2 variant F107A | | Descriptor: | CHLORIDE ION, Cyclooctat-9-en-7-ol synthase, MAGNESIUM ION | | Authors: | Driller, R, Janke, S, Fuchs, M, Warner, E, Mhashal, A.R, Major, D.T, Christmann, M, Brueck, T, Loll, B. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards a comprehensive understanding of the structural dynamics of a bacterial diterpene synthase during catalysis.
Nat Commun, 9, 2018
|
|
8UIR
 
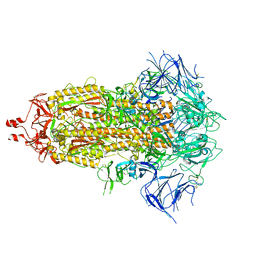 | |
1KRW
 
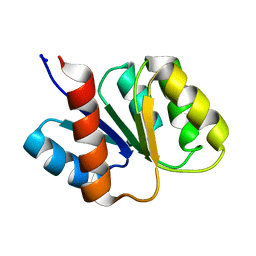 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | Descriptor: | NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
6JSG
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-chloropyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
8V0S
 
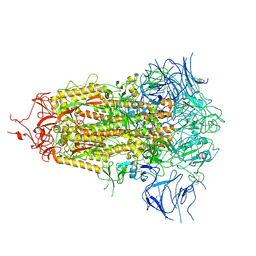 | |
6GMG
 
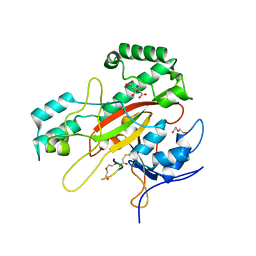 | | Structure of a glutamine donor mimicking inhibitory peptide shaped by the catalytic cleft of microbial transglutaminase | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE, ... | | Authors: | Schmelz, S, Juettner, N.E, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2018-05-25 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a glutamine donor mimicking inhibitory peptide shaped by the catalytic cleft of microbial transglutaminase.
FEBS J., 285, 2018
|
|
6JSZ
 
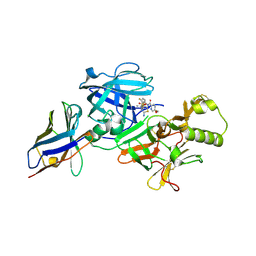 | | BACE2 xaperone complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 2, CHLORIDE ION, N-[3-[(5R)-3-azanyl-5-methyl-9,9-bis(oxidanylidene)-2,9$l^{6}-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluoranyl-phenyl]-5-(fluoranylmethoxy)pyrazine-2-carboxamide, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
8V0V
 
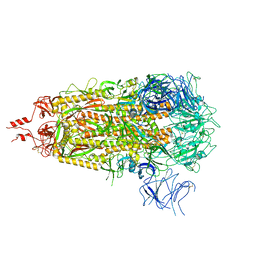 | |
6K3D
 
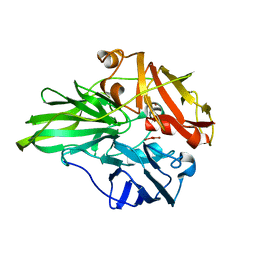 | | Structure of multicopper oxidase mutant | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, Multicopper oxidase | | Authors: | Sakuraba, H, Ohshida, T, Satomura, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Activity enhancement of multicopper oxidase from a hyperthermophile via directed evolution, and its application as the element of a high performance biocathode.
J.Biotechnol., 325, 2021
|
|
2FPG
 
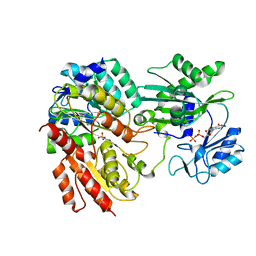 | |
3HT4
 
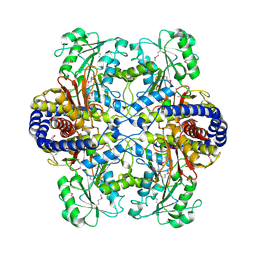 | | Crystal Structure of the Q81A77_BACCR Protein from Bacillus cereus. Northeast Structural Genomics Consortium Target BcR213 | | Descriptor: | Aluminum resistance protein | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Wang, H, Foote, E, Ciccosanti, C, Janjua, H, Xiao, R, Mao, L, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Q81A77_BACCR Protein from Bacillus cereus
To be Published
|
|
6JT4
 
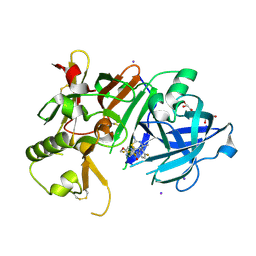 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
6GI6
 
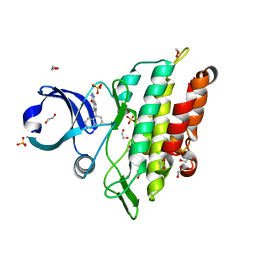 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 5-methyl core. | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-6-quinolin-5-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
3X2Q
 
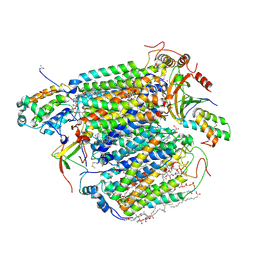 | | X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2014-12-26 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
8UKV
 
 | |
2OQ5
 
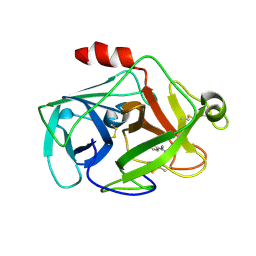 | | Crystal structure of DESC1, a new member of the type II transmembrane serine proteinases family | | Descriptor: | BENZAMIDINE, Transmembrane protease, serine 11E | | Authors: | Kyrieleis, O.J.P, Huber, R, Madison, E.L, Jacob, U. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of the catalytic domain of DESC1, a new member of the type II transmembrane serine proteinase family.
Febs J., 274, 2007
|
|
8UKF
 
 | |
4H8X
 
 | | Radiation damage study of lysozyme - 0.07 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1998 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6GQJ
 
 | | Crystal structure of human c-KIT kinase domain in complex with AZD3229-analogue (compound 18) | | Descriptor: | 2-[4-(6,7-dimethoxyquinazolin-4-yl)oxy-2-methoxy-phenyl]-~{N}-(1-propan-2-ylpyrazol-4-yl)ethanamide, Mast/stem cell growth factor receptor Kit | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
4H91
 
 | | Radiation damage study of lysozyme - 0.35 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2002 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2AXD
 
 | | solution structure of the theta subunit of escherichia coli DNA polymerase III in complex with the epsilon subunit | | Descriptor: | DNA polymerase III, theta subunit | | Authors: | Keniry, M.A, Park, A.Y, Owen, E.A, Hamdan, S.M, Pintacuda, G, Otting, G, Dixon, N.E. | | Deposit date: | 2005-09-05 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the theta subunit of Escherichia coli DNA polymerase III in complex with the epsilon subunit
J.Bacteriol., 188, 2006
|
|
8V0W
 
 | |
2P6P
 
 | |
