8ZI1
 
 | | Cryo-EM reveals transition states of the Acinetobacter baumannii F1-ATPase rotary subunits gamma and epsilon and novel compound targets - Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Le, K.C.M, Wong, C.F, Gruber, G. | | Deposit date: | 2024-05-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM reveals transition states of the Acinetobacter baumannii F 1 -ATPase rotary subunits gamma and epsilon , unveiling novel compound targets.
Faseb J., 38, 2024
|
|
8ZI2
 
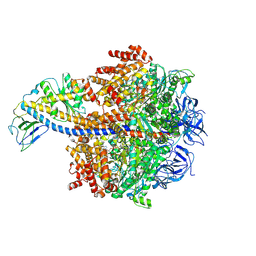 | | Cryo-EM reveals transition states of the Acinetobacter baumannii F1-ATPase rotary subunits gamma and epsilon and novel compound targets - Conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Le, K.C.M, Wong, C.F, Gruber, G. | | Deposit date: | 2024-05-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM reveals transition states of the Acinetobacter baumannii F 1 -ATPase rotary subunits gamma and epsilon , unveiling novel compound targets.
Faseb J., 38, 2024
|
|
5MSB
 
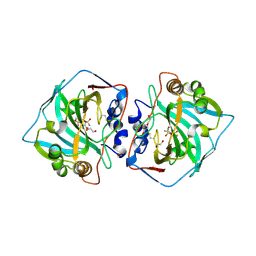 | | Crystal structure of human carbonic anhydrase isozyme XII with 2,3,5,6-tetrafluoro-4[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
9DCN
 
 | |
5J3Z
 
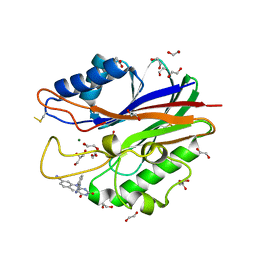 | | Crystal structure of m2hTDP2-CAT in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-10-[3-(1H-tetrazol-5-yl)phenyl]-2,3,4,10-tetrahydropyrimido[4,5-b]quinoline-8-carbonitrile, ACETATE ION, ... | | Authors: | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
8P0T
 
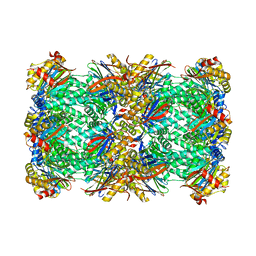 | | CryoEM structure of 20S Trichomonas vaginalis proteasome in complex with proteasome inhibitor CP-17 | | Descriptor: | Family T1, proteasome alpha subunit, threonine peptidase, ... | | Authors: | Silhan, J, Boura, E, Fajtova, P. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural elucidation of recombinant Trichomonas vaginalis 20S proteasome bound to covalent inhibitors.
Nat Commun, 15, 2024
|
|
9QQA
 
 | | Ternary complex of translating ribosome, NAC and NMT1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S19, ... | | Authors: | Echeverria, B, Jaskolowski, M, Scaiola, A, Ban, N. | | Deposit date: | 2025-03-31 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of cotranslational protein N-myristoylation in human cells.
Mol.Cell, 85, 2025
|
|
9QQB
 
 | | Quaternary complex of a translating ribosome, NAC, NMT1, and NatA | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S19, ... | | Authors: | Echeverria, B, Jaskolowski, M, Scaiola, A, Ban, N. | | Deposit date: | 2025-03-31 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Mechanism of cotranslational protein N-myristoylation in human cells.
Mol.Cell, 85, 2025
|
|
6EPB
 
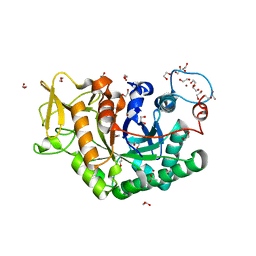 | | Structure of Chitinase 42 from Trichoderma harzianum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Endochitinase 42, ... | | Authors: | Ramirez-Escudero, M, Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Use of chitin and chitosan to produce new chitooligosaccharides by chitinase Chit42: enzymatic activity and structural basis of protein specificity.
Microb. Cell Fact., 17, 2018
|
|
9E8M
 
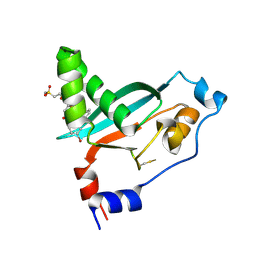 | | Covalent inhibitor VVD-442 bound to the RAS binding domain (RBD) of PI3Ka | | Descriptor: | 1-[(1P)-5-bromo-2'-chloro[1,1'-biphenyl]-2-sulfonyl]-4-fluoro-N-[(2S)-4-(methanesulfonyl)butan-2-yl]piperidine-4-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Bernard, S.M, Tamiya, J. | | Deposit date: | 2024-11-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.827 Å) | | Cite: | Covalent inhibitors of the RAS binding domain of PI3K alpha impair tumor growth driven by RAS and HER2.
Biorxiv, 2024
|
|
9QAV
 
 | | Human angiotensin-1 converting enzyme N-domain in complex with enalaprilat | | Descriptor: | 1,2-ETHANEDIOL, 1-((2S)-2-{[(1S)-1-CARBOXY-3-PHENYLPROPYL]AMINO}PROPANOYL)-L-PROLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gregory, K.S, Acharya, K.R. | | Deposit date: | 2025-02-28 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of domain-specific angiotensin I-converting enzyme inhibition by the antihypertensive drugs enalaprilat, ramiprilat, trandolaprilat, quinaprilat and perindoprilat.
Febs J., 2025
|
|
6HCV
 
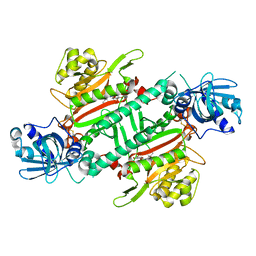 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with a chromone ligand | | Descriptor: | 6-fluoranyl-~{N}-[(1-oxidanylcyclohexyl)methyl]-4-oxidanylidene-chromene-2-carboxamide, Lysine--tRNA ligase | | Authors: | Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6TE6
 
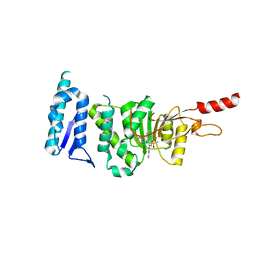 | | Crystal structure of Dot1L in complex with an inhibitor (compound 3). | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, ~{N}1-[(~{S})-(3-chlorophenyl)-pyridin-2-yl-methyl]-4-methylsulfonyl-~{N}2-pyrimidin-2-yl-benzene-1,2-diamine | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-11 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
8CKN
 
 | |
7QWO
 
 | | ATAD2 in complex with FragLite6 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1~{H}-pyridin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-25 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QXT
 
 | | ATAD2 in complex with FragLite10 | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-1,3-dihydro-2H-indol-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7R00
 
 | | ATAD2 in complex with FragLite33 | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-5-bromanyl-1-methyl-pyridin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-02-01 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
6YDM
 
 | | beta-phosphoglucomutase from Lactococcus lactis with citrate, tris and acetate bound | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
6Q7A
 
 | |
7ZZ9
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | 3-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(2-azanylethyl)amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]-3-oxidanylidene-propanoic acid, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
8A9V
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-2-azanyl-ethanesulfonamide | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
8Z48
 
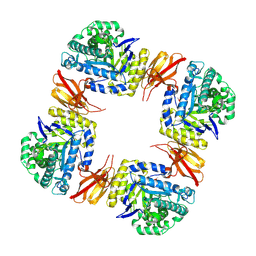 | | Beta-galactosidase from Bacteroides xylanisolvens (complex with methyl beta-galactopyranose) | | Descriptor: | Beta-galactosidase, methyl beta-D-galactopyranoside | | Authors: | Nakajima, M, Motouchi, S, Kobayashi, K. | | Deposit date: | 2024-04-16 | | Release date: | 2025-01-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and function of a beta-1,2-galactosidase from Bacteroides xylanisolvens, an intestinal bacterium.
Commun Biol, 8, 2025
|
|
8R48
 
 | |
8R4G
 
 | |
