2FUJ
 
 | |
5FN5
 
 | | Cryo-EM structure of gamma secretase in class 3 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FLG
 
 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW)from Bacillus subtilis in complex with AMPPNP | | Descriptor: | 6-CARBOXYHEXANOATE--COA LIGASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
9FVO
 
 | | The HIV protease inhibitor amprenavir binding to the active site of Cryphonectria parasitica endothiapepsin | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Endothiapepsin, ... | | Authors: | Falke, S, Senst, J.M, Guenther, S, Meents, A. | | Deposit date: | 2024-06-27 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The HIV protease inhibitor amprenavir binding to the active site of Cryphonectria parasitica endothiapepsin
To Be Published
|
|
5FN2
 
 | | Cryo-EM structure of gamma secretase in complex with a drug DAPT | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN3
 
 | | Cryo-EM structure of gamma secretase in class 1 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN4
 
 | | Cryo-EM structure of gamma secretase in class 2 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
2FA5
 
 | | The crystal structure of an unliganded multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestris | | Descriptor: | CHLORIDE ION, transcriptional regulator marR/emrR family | | Authors: | Chin, K.H, Tu, Z.L, Li, J.N, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2005-12-06 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of XC1739: a putative multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestris at 1.8 A resolution
Proteins, 65, 2006
|
|
6C06
 
 | | Mycobacterium tuberculosis RNAP Holo/RbpA/Fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
2GM2
 
 | | NMR structure of Xanthomonas campestris XCC1710: Northeast Structural Genomics Consortium target XcR35 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Xiao, R, Wang, D.Y, Ma, L.C, Ciano, M, Montelione, G.T, Ramelot, T.A, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Xanthomonas campestris XCC1710 protein
To be Published
|
|
1HY9
 
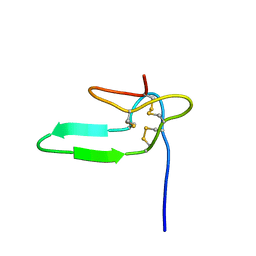 | | COCAINE AND AMPHETAMINE REGULATED TRANSCRIPT | | Descriptor: | COCAINE AND AMPHETAMINE REGULATED TRANSCRIPT PROTEIN | | Authors: | Ludvigsen, S, Thim, L, Blom, A.M, Wulff, B.S. | | Deposit date: | 2001-01-18 | | Release date: | 2001-08-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the satiety factor, CART, reveals new functionality of a well-known fold.
Biochemistry, 40, 2001
|
|
2HWN
 
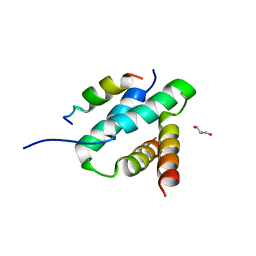 | |
2D28
 
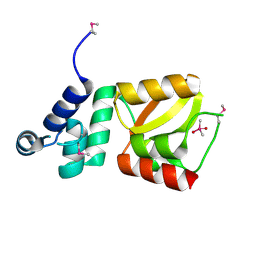 | | Structure of the N-terminal domain of XpsE (crystal form P43212) | | Descriptor: | CACODYLATE ION, type II secretion ATPase XpsE | | Authors: | Chen, Y, Shiue, S.-J, Huang, C.-W, Chang, J.-L, Chien, Y.-L, Hu, N.-T, Chan, N.-L. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of the XpsE N-Terminal Domain, an Essential Component of the Xanthomonas campestris Type II Secretion System
J.Biol.Chem., 280, 2005
|
|
5UAH
 
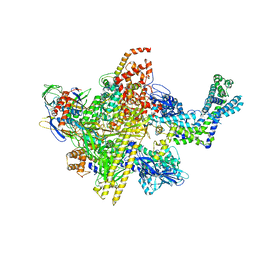 | | Escherichia coli RNA polymerase and Rifampin complex, RpoB D516V mutant | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
5UAL
 
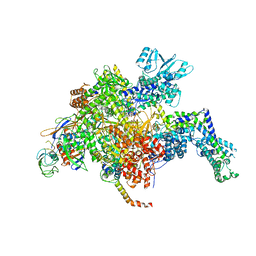 | | Escherichia coli RNA polymerase and Rifampin complex, RpoB S531L mutant | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.887 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
5UAC
 
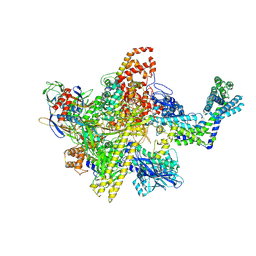 | | Escherichia coli RNA polymerase and Rifampin complex, wild-type | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
6WED
 
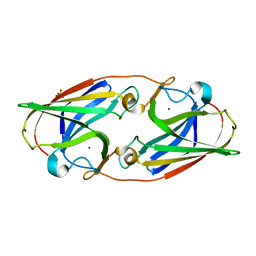 | |
6WEE
 
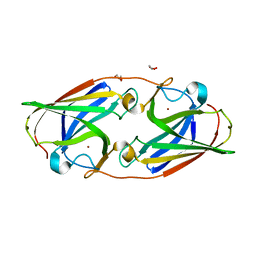 | | Copper-bound M88I variant of Campylobacter jejuni P19 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chan, A.C, Murphy, M.E. | | Deposit date: | 2020-04-02 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A copper site is required for iron transport by the periplasmic proteins P19 and FetP.
Metallomics, 12, 2020
|
|
6WEF
 
 | |
4ZOW
 
 | | Crystal structure of E. coli multidrug transporter MdfA in complex with chloramphenicol | | Descriptor: | CHLORAMPHENICOL, Multidrug transporter MdfA | | Authors: | Zhang, X.C, Heng, J, Zhao, Y, Wang, X. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate-bound structure of the E. coli multidrug resistance transporter MdfA
Cell Res., 25, 2015
|
|
2AV6
 
 | |
2AZD
 
 | |
2WD3
 
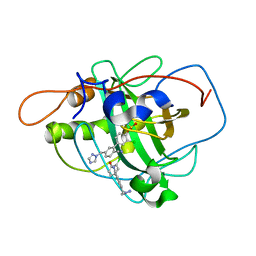 | | Highly Potent First Examples of Dual Aromatase-Steroid Sulfatase Inhibitors based on a Biphenyl Template | | Descriptor: | 3-CHLORO-2'-CYANO-5'-(1H-1,2,4-TRIAZOL-1-YLMETHYL)BIPHENYL-4-YL SULFAMATE, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Woo, L.W.L, Jackson, T, Putey, A, Cozier, G, Leonard, P, Acharya, K.R, Chander, S.K, Purohit, A, Reed, M.J, Potter, B.V.L. | | Deposit date: | 2009-03-19 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly Potent First Examples of Dual Aromatase-Steroid Sulfatase Inhibitors Based on a Biphenyl Template.
J.Med.Chem., 53, 2010
|
|
4ZER
 
 | | Crystal structure of the Onc112 antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16s ribosomal RNA, 23s ribosomal RNA, ... | | Authors: | Seefeldt, A.C, Nguyen, F, Antunes, S, Perebaskine, N, Graf, M, Arenz, S, Inampudi, K.K, Douat, C, Guichard, G, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-04-20 | | Release date: | 2015-05-20 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4TNJ
 
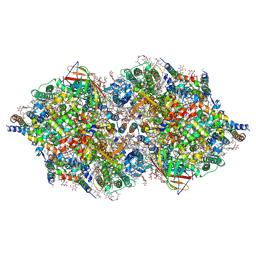 | | RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 4.5 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
