1OEC
 
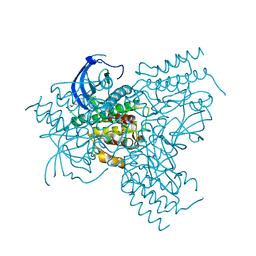 | | FGFr2 kinase domain | | Descriptor: | 4-ARYL-2-PHENYLAMINO PYRIMIDINE, FIBROBLAST GROWTH FACTOR RECEPTOR 2, SULFATE ION | | Authors: | Ceska, T.A, Owens, R, Doyle, C, Hamlyn, P, Crabbe, T, Moffat, D, Davis, J, Martin, R, Perry, M.J. | | Deposit date: | 2003-03-24 | | Release date: | 2004-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Fgfr2 Tyrosine Kinase Domain in Complex with 4-Aryl-2-Phenylamino Pyrimidine Angiogenesis Inhibitors
To be Published
|
|
1E7F
 
 | |
1E7H
 
 | |
1O46
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU90395. | | Descriptor: | 2-{4-[2-ACETYLAMINO-2-(1-BIPHENYL-4-YLMETHYL-2-OXO-AZEPAN-3-YLCARBAMOYL)-ETHYL]-2-METHOXYCARBONYL-PHENYL}-2-FLUORO-MALONIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1E7A
 
 | |
1E8D
 
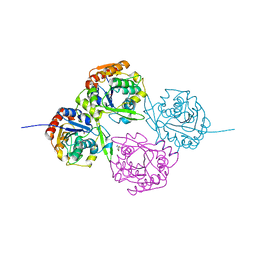 | | MECHANISTIC ASPECTS OF CYANOGENESIS FROM ACTIVE SITE MUTANT SER80ALA OF HYDROXYNITRILE LYASE FROM MANIHOT ESCULENTA IN COMPLEX WITH ACETONE CYANOHYDRIN | | Descriptor: | 2-HYDROXY-2-METHYLPROPANENITRILE, HYDROXYNITRILE LYASE | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Wajant, H, Effenberger, F. | | Deposit date: | 2000-09-19 | | Release date: | 2001-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Aspects of Cyanogenesis from Active-Site Mutant Ser80Ala of Hydroxynitrile Lyase from Manihot Esculenta in Complex with Acetone Cyanohydrin.
Protein Sci., 10, 2001
|
|
1E78
 
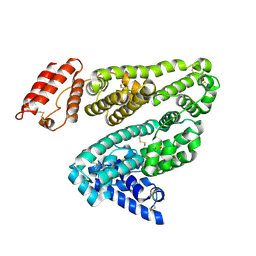 | |
1E7B
 
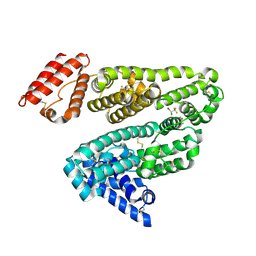 | | Crystal structure of human serum albumin complexed with the general anesthetic halothane | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, SERUM ALBUMIN | | Authors: | Bhattacharya, A.A, Curry, S, Franks, N.P. | | Deposit date: | 2000-08-26 | | Release date: | 2001-01-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Binding of the General Anesthetics Propofol and Halothane to Human Serum Albumin. High Resolution Crystal Structures
J.Biol.Chem., 275, 2000
|
|
1OCT
 
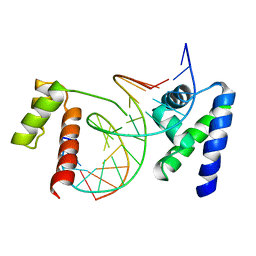 | | CRYSTAL STRUCTURE OF THE OCT-1 POU DOMAIN BOUND TO AN OCTAMER SITE: DNA RECOGNITION WITH TETHERED DNA-BINDING MODULES | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*TP*AP*TP*TP*TP*GP*CP*AP*TP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*CP*AP*AP*AP*TP*AP*AP*GP*G)-3'), PROTEIN (OCT-1 POU DOMAIN) | | Authors: | Klemm, J.D, Rould, M.A, Aurora, R, Herr, W, Pabo, C.O. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules.
Cell(Cambridge,Mass.), 77, 1994
|
|
1E7G
 
 | |
1IDN
 
 | | MAC-1 I DOMAIN METAL FREE | | Descriptor: | CD11B | | Authors: | Baldwin, E.T. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cation binding to the integrin CD11b I domain and activation model assessment
Structure, 6, 1998
|
|
1OEL
 
 | |
1OBM
 
 | |
1OBS
 
 | | STRUCTURE OF RICIN A CHAIN MUTANT | | Descriptor: | RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-25 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
1OBT
 
 | | STRUCTURE OF RICIN A CHAIN MUTANT, COMPLEX WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-22 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
1NUC
 
 | | STAPHYLOCOCCAL NUCLEASE, V23C VARIANT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1997-02-15 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mobile unnatural amino acid side chains in the core of staphylococcal nuclease.
Protein Sci., 5, 1996
|
|
1NWO
 
 | |
1NOV
 
 | | NODAMURA VIRUS | | Descriptor: | NODAMURA VIRUS COAT PROTEINS | | Authors: | Natarajan, P, Johnson, J.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Resolution of space-group ambiguity and structure determination of nodamura virus to 3.3 A resolution from pseudo-R32 (monoclinic) crystals.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1NWP
 
 | |
1O9C
 
 | | Structural view of a fungal toxin acting on a 14-3-3 regulatory complex | | Descriptor: | 14-3-3-LIKE PROTEIN C, CHLORIDE ION, CITRATE ANION | | Authors: | Wurtele, M, Jelich-Ottmann, C, Wittinghofer, A, Oecking, C. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex
Embo J., 22, 2003
|
|
1O99
 
 | | CRYSTAL STRUCTURE OF THE S62A MUTANT OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | Descriptor: | 2,3-BISPHOSPHOGLYCERATE-INDEPENDENT PHOSPHOGLYCERATE MUTASE, 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, ... | | Authors: | Rigden, D.J, Lamani, E, Littlejohn, J.E, Jedrzejas, M.J. | | Deposit date: | 2002-12-11 | | Release date: | 2002-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Insights Into the Catalytic Mechanism of Cofactor-Independent Phosphoglycerate Mutase from X-Ray Crystallography, Simulated Dynamics and Molecular Modeling
J.Mol.Biol., 328, 2003
|
|
1O8M
 
 | |
1OE0
 
 | | CRYSTAL STRUCTURE OF DROSOPHILA DEOXYRIBONUCLEOSIDE KINASE IN COMPLEX WITH DTTP | | Descriptor: | DEOXYRIBONUCLEOSIDE KINASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-17 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway:Studies of the Drosophila Deoxyribonucleoside Kinase
Biochemistry, 42, 2003
|
|
1O8D
 
 | |
1O90
 
 | | Methionine Adenosyltransferase complexed with a L-methionine analogue | | Descriptor: | (2S,4S)-2-AMINO-4,5-EPOXIPENTANOIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
