5L6H
 
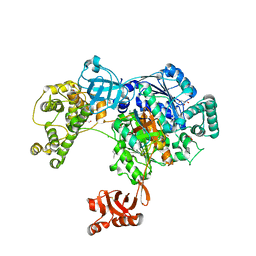 | | Uba1 in complex with Ub-ABPA3 covalent adduct | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
5L6I
 
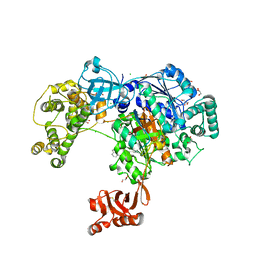 | | Uba1 in complex with Ub-MLN4924 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
5L6J
 
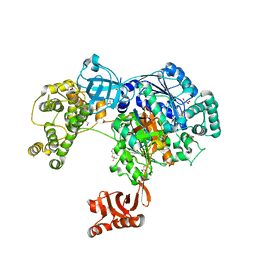 | | Uba1 in complex with Ub-MLN7243 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
2IY5
 
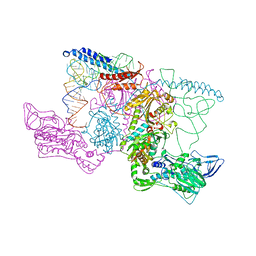 | | PHENYLALANYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS complexed with tRNA and a phenylalanyl-adenylate analog | | Descriptor: | ADENOSINE-5'-[PHENYLALANINOL-PHOSPHATE], MAGNESIUM ION, PHENYLALANYL-TRNA SYNTHETASE ALPHA CHAIN, ... | | Authors: | Moor, N, Kotik-Kogan, O, Tworowski, D, Sukhanova, M, Safro, M. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of the ternary complex of phenylalanyl-tRNA synthetase with tRNAPhe and a phenylalanyl-adenylate analogue reveals a conformational switch of the CCA end.
Biochemistry, 45, 2006
|
|
5NDG
 
 | | Crystal structure of geneticin (G418) bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3PCO
 
 | | crystal structure of E. coli phenylalanine-tRNA synthetase complexed with phenylalanine and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHENYLALANINE, Phenylalanyl-tRNA synthetase, ... | | Authors: | Mermershtain, I, Finarov, I, Klipcan, L, Kessler, N, Rozenberg, H, Safro, M.G. | | Deposit date: | 2010-10-21 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Idiosyncrasy and identity in the prokaryotic phe-system: crystal structure of E. coli phenylalanyl-tRNA synthetase complexed with phenylalanine and AMP.
Protein Sci., 20, 2011
|
|
8ALZ
 
 | | Cryo-EM structure of ASCC3 in complex with ASC1 | | Descriptor: | Activating signal cointegrator 1, Activating signal cointegrator 1 complex subunit 3, ZINC ION | | Authors: | Jia, J, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of ASCC3 in complex with ASC1
Nat Commun, 2023
|
|
6P5J
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 2) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P59
 
 | | Crystal structure of SIVrcm Vif-CBFbeta-ELOB-ELOC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Core-binding factor subunit beta, Elongin-B, ... | | Authors: | Binning, J.M, Chesarino, N.M, Emerman, M, Gross, J.D. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.942214 Å) | | Cite: | Structural Basis for a Species-Specific Determinant of an SIV Vif Protein toward Hominid APOBEC3G Antagonism.
Cell Host Microbe, 26, 2019
|
|
3VX7
 
 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | Descriptor: | E1, E2 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VX6
 
 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1KSZ
 
 | | ENTRAPMENT OF 6-THIOPHOSPHORYL-IMP IN THE ACTIVE SITE OF CRYSTALLINE ADENYLOSUCCINATE SYNTHETASE FROM ESCHERICHIA COLI, DATA COLLECTED AT 298K | | Descriptor: | 2-DEAZO-6-THIOPHOSPHATE GUANOSINE-5'-MONOPHOSPHATE, ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Poland, B.W, Bruns, C.A, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1997-01-14 | | Release date: | 1997-10-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Entrapment of 6-thiophosphoryl-IMP in the active site of crystalline adenylosuccinate synthetase from Escherichia coli.
J.Biol.Chem., 272, 1997
|
|
8RO0
 
 | | Structure of the C. elegans Intron Lariat Spliceosome primed for disassembly (ILS') | | Descriptor: | Cell division cycle 5-like protein, Coiled-coil domain-containing protein 12, GCF C-terminal domain-containing protein, ... | | Authors: | Vorlaender, M.K, Rothe, P, Plaschka, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8C2S
 
 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 1). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8CA3
 
 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 2). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
1U0F
 
 | | Crystal structure of mouse phosphoglucose isomerase in complex with glucose 6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, GLUCOSE-6-PHOSPHATE, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
7W3K
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD4_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3A
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED4_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W37
 
 | | Structure of USP14-bound human 26S proteasome in state EA1_UBL | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3F
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED1_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W38
 
 | | Structure of USP14-bound human 26S proteasome in state EA2.0_UBL | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3I
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SB_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3M
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD5_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
1U0G
 
 | | Crystal structure of mouse phosphoglucose isomerase in complex with erythrose 4-phosphate | | Descriptor: | BETA-MERCAPTOETHANOL, ERYTHOSE-4-PHOSPHATE, GLYCEROL, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0E
 
 | | Crystal structure of mouse phosphoglucose isomerase | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
