1YCS
 
 | | P53-53BP2 COMPLEX | | Descriptor: | 53BP2, P53, ZINC ION | | Authors: | Gorina, S, Pavletich, N.P. | | Deposit date: | 1996-09-30 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2.
Science, 274, 1996
|
|
2ATA
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex II) | | Descriptor: | 5'-D(*AP*AP*GP*GP*CP*AP*TP*GP*CP*CP*TP*T)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-08-24 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
5LGY
 
 | |
6SL6
 
 | | p53 charged core | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, ZINC ION | | Authors: | Gallardo, R, Langenberg, T, Schymkowitz, J, Rousseau, F, Ulens, C. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Thermodynamic and Evolutionary Coupling between the Native and Amyloid State of Globular Proteins.
Cell Rep, 31, 2020
|
|
5MCU
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LHG2) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5MCV
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LWC1) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cellular tumor antigen p53, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5MF7
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-GADD45) | | Descriptor: | Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, DNA, ... | | Authors: | Rozenberg, H, Diskin-Posner, Y, Golovenko, D, Shakked, Z. | | Deposit date: | 2016-11-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5MCT
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LHG1) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4CHA
 
 | |
5MG7
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-p53R2) | | Descriptor: | Cellular tumor antigen p53, DNA, ZINC ION | | Authors: | Rozenberg, H, Braeuning, B, Golovenko, D, Shakked, Z. | | Deposit date: | 2016-11-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5MCW
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LWC2) | | Descriptor: | Cellular tumor antigen p53, DNA, FORMYL GROUP, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
7JRX
 
 | | Crystal structure of the R64F mutant of Bauhinia Bauhinioides complexed with Bovine Chymotrypsin | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-13 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6UE7
 
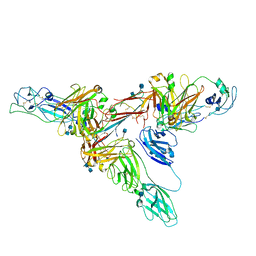 | | Structure of dimeric sIgA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
8J8N
 
 | |
2YBG
 
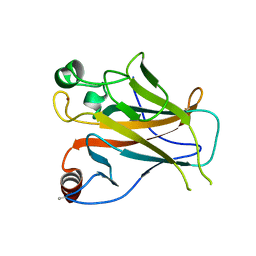 | | Structure of Lys120-acetylated p53 core domain | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Arbely, E, Allen, M.D, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2011-03-08 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acetylation of Lysine 120 of P53 Endows DNA- Binding Specificity at Effective Physiological Salt Concentration.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3T62
 
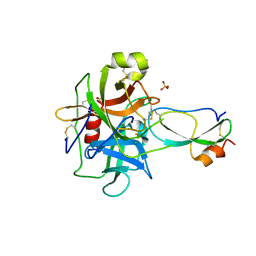 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean Sea anemone Stichodactyla helianthus in complex with bovine chymotrypsin | | Descriptor: | Chymotrypsinogen A, Kunitz-type proteinase inhibitor SHPI-1, SULFATE ION | | Authors: | Garcia-Fernandez, R, Dominguez, R, Oberthuer, D, Pons, T, Gonzalez-Gonzalez, Y, Chavez, M.A, Betzel, C, Redecke, L. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into chymotrypsin inhibition by the Kunitz-type inhibitor-1 from the marine invertebrate Stichodactyla helianthus
To be Published
|
|
2Y6T
 
 | | Molecular Recognition of Chymotrypsin by the Serine Protease Inhibitor Ecotin from Yersinia pestis | | Descriptor: | CHYMOTRYPSINOGEN A, ECOTIN, SULFATE ION | | Authors: | Clark, E.A, Walker, N, Ford, D.C, Cooper, I.A, Oyston, P.C.F, Acharya, K.R. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular Recognition of Chymotrypsin by the Serine Protease Inhibitor Ecotin from Yersinia Pestis.
J.Biol.Chem., 286, 2011
|
|
8GCH
 
 | |
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Wei, H, Guo, M, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
