6CR1
 
 | | adalimumab EFab | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heavy chain of adalimumab EFab (VH-IgE CH2), Light chain of adalimumab EFab (VL-IgE CH2), ... | | Authors: | Arndt, J.W. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | EFab domain substitution as a solution to the light-chain pairing problem of bispecific antibodies.
MAbs, 10, 2018
|
|
6UX5
 
 | |
7VKE
 
 | | Crystal structure of human CD38 ECD in complex with UniDab(TM) F11A | | Descriptor: | 1,2-ETHANEDIOL, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, CHLORIDE ION, ... | | Authors: | Schooten, W.V, Schellenberger, U, Ugamraj, H.S, Manicka, S, Bijpuria, S, Gondu, R.K. | | Deposit date: | 2021-09-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TNB-738, a biparatopic antibody, boosts intracellular NAD+ by inhibiting CD38 ecto-enzyme activity.
Mabs, 14, 2022
|
|
6X3I
 
 | | NNAS Fc mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-mannopyranose | | Authors: | Wei, R, Zhou, Y.F. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Engineered Fc-glycosylation switch to eliminate antibody effector function.
Mabs, 12, 2020
|
|
6R35
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PAO1 in complex with lewis x tetrasaccharide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Lepsik, M, Sommer, R, Kuhaudomlarp, S, Lelimousin, M, Varrot, A, Titz, A, Imberty, A. | | Deposit date: | 2019-03-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induction of rare conformation of oligosaccharide by binding to calcium-dependent bacterial lectin: X-ray crystallography and modelling study.
Eur.J.Med.Chem., 177, 2019
|
|
6XSJ
 
 | |
2PZ0
 
 | | Crystal structure of Glycerophosphodiester Phosphodiesterase (GDPD) from T. tengcongensis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycerophosphoryl diester phosphodiesterase | | Authors: | Shi, L, Liu, J.F, An, X.M, Liang, D.C. | | Deposit date: | 2007-05-17 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of glycerophosphodiester phosphodiesterase (GDPD) from Thermoanaerobacter tengcongensis, a metal ion-dependent enzyme: insight into the catalytic mechanism.
Proteins, 72, 2008
|
|
6NOU
 
 | | An scFv derived from ixekizumab | | Descriptor: | GLYCEROL, scFv derived from ixekizumab | | Authors: | Durbin, J.D, Clawson, D.K, Lu, F, Tian, Y, Lu, J, Atwell, S. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease.
Mabs, 11, 2019
|
|
6OEC
 
 | | Yeast Spc42 Trimeric Coiled-Coil Amino Acids 181-211 fused to PDB: 3H5I | | Descriptor: | CALCIUM ION, Response regulator/sensory box protein/GGDEF domain protein,Spindle pole body component SPC42 | | Authors: | Drennan, A.C, Shivaani, K, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jasperson, S.L, Rayment, I. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
8I5P
 
 | |
8I5Q
 
 | |
8I5T
 
 | |
8I5U
 
 | |
6XSV
 
 | |
8I5S
 
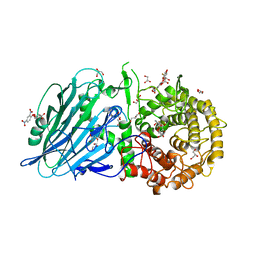 | | Crystal structure of TxGH116 D593N acid/base mutant from Thermoanaerobacterium xylanolyticum with 2-deoxy-2-fluoroglucoside | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reaction Mechanism of Glycoside Hydrolase Family 116 Utilizes Perpendicular Protonation.
Acs Catalysis, 13, 2023
|
|
5JPN
 
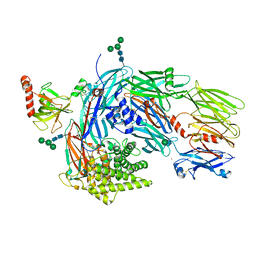 | | Structure of human complement C4 rebuilt using iMDFF | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C4-A, ... | | Authors: | Croll, T.I, Andersen, G.R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Re-evaluation of low-resolution crystal structures via interactive molecular-dynamics flexible fitting (iMDFF): a case study in complement C4.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6NQY
 
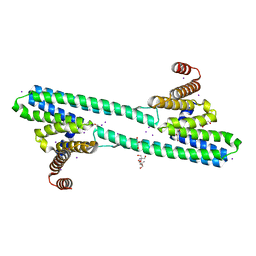 | | Flagellar protein FcpA from Leptospira biflexa / ab-centered monoclinic form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Flagellar coiling protein A, ... | | Authors: | Mechaly, A, Larrieux, N, Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete.
Elife, 9, 2020
|
|
6NQX
 
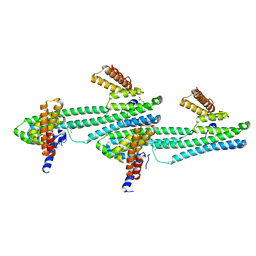 | | Flagellar protein FcpA from Leptospira biflexa / primitive monoclinic form | | Descriptor: | Flagellar coiling protein A, GLYCEROL | | Authors: | Mechaly, A, Larrieux, N, Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete
Elife, 9, 2020
|
|
6NQW
 
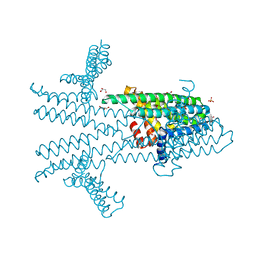 | | Flagellar protein FcpA from Leptospira biflexa - hexagonal form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Flagellar coiling protein A, GLYCEROL, ... | | Authors: | San Martin, F, Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete
Elife, 9, 2020
|
|
3FUB
 
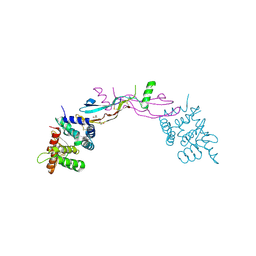 | | Crystal structure of GDNF-GFRalpha1 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parkash, V, Goldman, A. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Comparison of GFL-GFRalpha complexes: further evidence relating GFL bend angle to RET signalling
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6TOW
 
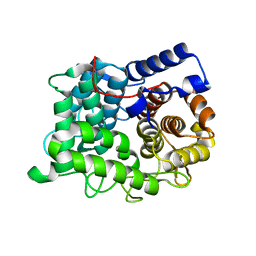 | |
6TRH
 
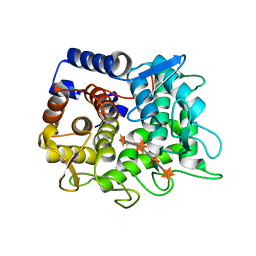 | |
6TO0
 
 | |
5JTW
 
 | | Crystal structure of complement C4b re-refined using iMDFF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C4-A | | Authors: | Croll, T.I, Andersen, G.R. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Re-evaluation of low-resolution crystal structures via interactive molecular-dynamics flexible fitting (iMDFF): a case study in complement C4.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3SI2
 
 | | Structure of glycosylated murine glutaminyl cyclase in presence of the inhibitor PQ50 (PDBD150) | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Parthier, C, Carrillo, D, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
