1STD
 
 | |
1STE
 
 | |
1STF
 
 | |
1STG
 
 | | TWO DISTINCTLY DIFFERENT METAL BINDING MODES ARE SEEN IN X-RAY CRYSTAL STRUCTURES OF STAPHYLOCOCCAL NUCLEASE-COBALT(II)-NUCLEOTIDE COMPLEXES | | Descriptor: | COBALT (II) ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Loll, P.J, Quirk, S, Lattman, E.E. | | Deposit date: | 1994-10-27 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structures of staphylococcal nuclease complexed with the competitive inhibitor cobalt(II) and nucleotide.
Biochemistry, 34, 1995
|
|
1STH
 
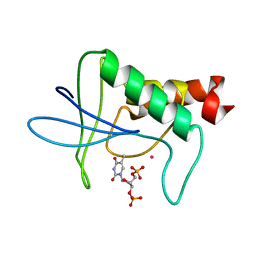 | | TWO DISTINCTLY DIFFERENT METAL BINDING MODES ARE SEEN IN X-RAY CRYSTAL STRUCTURES OF STAPHYLOCOCCAL NUCLEASE-COBALT(II)-NUCLEOTIDE COMPLEXES | | Descriptor: | COBALT (II) ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Loll, P.J, Quirk, S, Lattman, E.E. | | Deposit date: | 1994-10-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structures of staphylococcal nuclease complexed with the competitive inhibitor cobalt(II) and nucleotide.
Biochemistry, 34, 1995
|
|
1STM
 
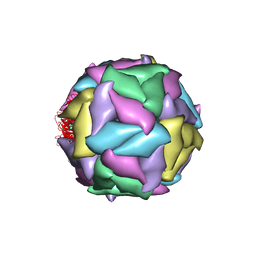 | | SATELLITE PANICUM MOSAIC VIRUS | | Descriptor: | SATELLITE PANICUM MOSAIC VIRUS | | Authors: | Ban, N, McPherson, A. | | Deposit date: | 1995-07-12 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of satellite panicum mosaic virus at 1.9 A resolution.
Nat.Struct.Biol., 2, 1995
|
|
1STN
 
 | |
1STO
 
 | |
1STP
 
 | |
1STQ
 
 | | Cyrstal Structure of Cytochrome c Peroxidase Mutant: CcPK2M3 | | Descriptor: | Cytochrome c peroxidase, mitochondrial, POTASSIUM ION, ... | | Authors: | Barrows, T.P, Bhaskar, B, Poulos, T.L. | | Deposit date: | 2004-03-25 | | Release date: | 2004-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Electrostatic control of the tryptophan radical in cytochrome c peroxidase.
Biochemistry, 43, 2004
|
|
1STR
 
 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE AC-CHPQNT-NH2 DIMER | | Descriptor: | AC-CHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
1STS
 
 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE FCHPQNT-NH2 DIMER | | Descriptor: | FCHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
1STU
 
 | | DOUBLE STRANDED RNA BINDING DOMAIN | | Descriptor: | MATERNAL EFFECT PROTEIN STAUFEN | | Authors: | Bycroft, M. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5.
EMBO J., 14, 1995
|
|
1STX
 
 | |
1STY
 
 | |
1STZ
 
 | | Crystal structure of a hypothetical protein at 2.2 A resolution | | Descriptor: | Heat-inducible transcription repressor hrcA homolog | | Authors: | Liu, J, Adams, P.D, Shin, D.-H, Huang, C, Yokota, H, Jancarik, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a heat-inducible transcriptional repressor HrcA from Thermotoga maritima: structural insight into DNA binding and dimerization.
J.Mol.Biol., 350, 2005
|
|
1SU0
 
 | | Crystal structure of a hypothetical protein at 2.3 A resolution | | Descriptor: | NifU like protein IscU, ZINC ION | | Authors: | Liu, J, Oganesyan, N, Shin, D.-H, Jancarik, J, Pufan, R, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an iron-sulfur cluster assembly protein IscU in a zinc-bound form.
Proteins, 59, 2005
|
|
1SU1
 
 | | Structural and biochemical characterization of Yfce, a phosphoesterase from E. coli | | Descriptor: | Hypothetical protein yfcE, SULFATE ION, ZINC ION | | Authors: | Miller, D.J, Shuvalova, L, Evdokimova, E, Savchenko, A, Yakunin, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biochemical characterization of a novel Mn2+-dependent phosphodiesterase encoded by the yfcE gene.
Protein Sci., 16, 2007
|
|
1SU2
 
 | | CRYSTAL STRUCTURE OF THE NUDIX HYDROLASE DR1025 IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MutT/nudix family protein | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1SU3
 
 | | X-ray structure of human proMMP-1: New insights into collagenase action | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jozic, D, Bourenkov, G, Lim, N.H, Nagase, H, Bode, W, Maskos, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-26 | | Release date: | 2004-12-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of human proMMP-1: new insights into procollagenase activation and collagen binding.
J.Biol.Chem., 280, 2005
|
|
1SU4
 
 | | Crystal structure of calcium ATPase with two bound calcium ions | | Descriptor: | CALCIUM ION, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Toyoshima, C, Nakasako, M, Nomura, H, Ogawa, H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution
Nature, 405, 2000
|
|
1SU5
 
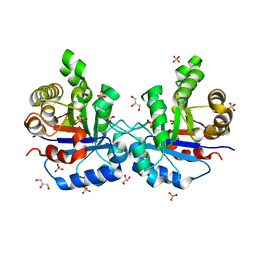 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SU6
 
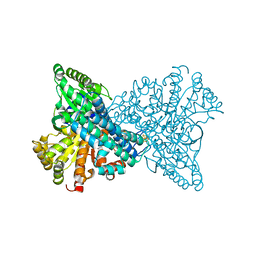 | | Carbon monoxide dehydrogenase from Carboxydothermus hydrogenoformans: CO reduced state | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
1SU7
 
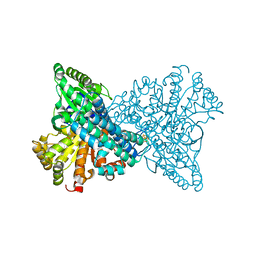 | | Carbon Monoxide Dehydrogenase from Carboxydothermus hydrogenoformans- DTT reduced state | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
1SU8
 
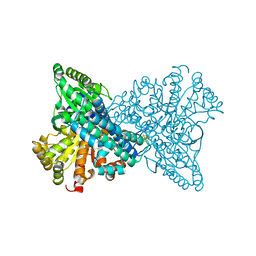 | | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
