1XZD
 
 | |
1A8Y
 
 | | CRYSTAL STRUCTURE OF CALSEQUESTRIN FROM RABBIT SKELETAL MUSCLE SARCOPLASMIC RETICULUM AT 2.4 A RESOLUTION | | Descriptor: | CALSEQUESTRIN | | Authors: | Wang, S, Trumble, W.R, Liao, H, Wesson, C.R, Dunker, A.K, Kang, C. | | Deposit date: | 1998-03-31 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum.
Nat.Struct.Biol., 5, 1998
|
|
1V8W
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E82Q mutant, complexed with SO4 and Zn | | Descriptor: | ADP-ribose pyrophosphatase, SULFATE ION, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
2D2I
 
 | | Crystal Structure of NADP-Dependent Glyceraldehyde-3-Phosphate Dehydrogenase from Synechococcus Sp. complexed with Nadp+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, glyceraldehyde 3-phosphate dehydrogenase | | Authors: | Kitatani, T, Nakamura, Y, Wada, K, Kinoshita, T, Tamoi, M, Shigeoka, S, Tada, T. | | Deposit date: | 2005-09-09 | | Release date: | 2006-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Synechococcus PCC7942 complexed with NADP
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1V3C
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with NEU5AC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1XZF
 
 | |
1A76
 
 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MANGANESE (II) ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
1UN6
 
 | | THE CRYSTAL STRUCTURE OF A ZINC FINGER - RNA COMPLEX REVEALS TWO MODES OF MOLECULAR RECOGNITION | | Descriptor: | 5S RIBOSOMAL RNA, MAGNESIUM ION, TRANSCRIPTION FACTOR IIIA, ... | | Authors: | Lu, D, Searles, M.A, Klug, A. | | Deposit date: | 2003-09-04 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Zinc-Finger-RNA Complex Reveals Two Modes of Molecular Recognition
Nature, 426, 2003
|
|
1XKJ
 
 | | BACTERIAL LUCIFERASE BETA2 HOMODIMER | | Descriptor: | BETA2 LUCIFERASE | | Authors: | Tanner, J.J, Krause, K.L. | | Deposit date: | 1996-10-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial luciferase beta 2 homodimer: implications for flavin binding.
Biochemistry, 36, 1997
|
|
1XSM
 
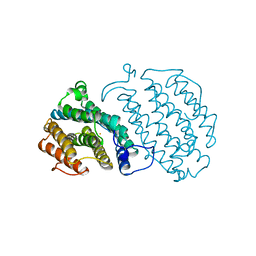 | | PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE FROM MOUSE | | Descriptor: | FE (III) ION, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Kauppi, B, Nielsen, B.N, Ramaswamy, S, Kjoller-Larsen, I, Thelander, M, Thelander, L, Eklund, H. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2.
J.Mol.Biol., 262, 1996
|
|
1UYT
 
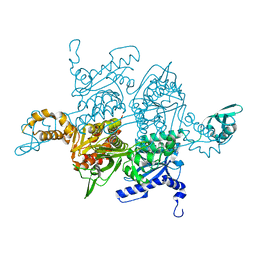 | | Acetyl-CoA carboxylase carboxyltransferase domain | | Descriptor: | ACETYL-COA CARBOXYLASE | | Authors: | Zhang, H, Tweel, B, Tong, L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Inhibition of the Carboxyltransferase Domain of Acetyl-Coenzyme-A Carboxylase by Haloxyfop and Diclofop.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XWL
 
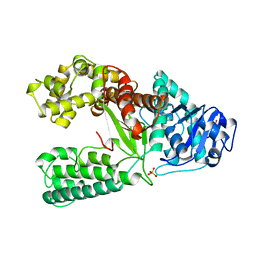 | |
1XGO
 
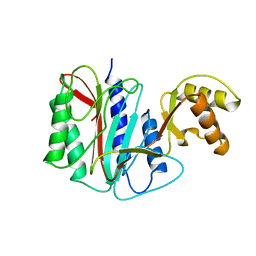 | |
1XGN
 
 | |
1UVU
 
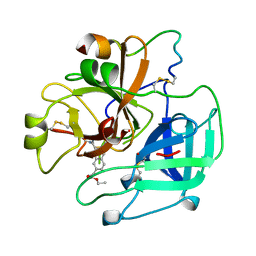 | | BOVINE THROMBIN--BM12.1700 COMPLEX | | Descriptor: | 3-(7-DIAMINOMETHYL-NAPHTHALEN-2-YL)-PROPIONIC ACID ETHYL ESTER, THROMBIN | | Authors: | Engh, R.A, Huber, R. | | Deposit date: | 1996-10-16 | | Release date: | 1997-11-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme flexibility, solvent and 'weak' interactions characterize thrombin-ligand interactions: implications for drug design.
Structure, 4, 1996
|
|
1XAC
 
 | |
1XYN
 
 | |
1XAD
 
 | |
1XRC
 
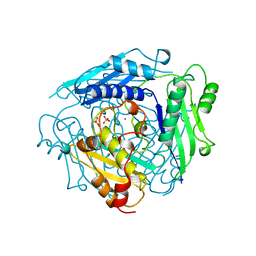 | | CRYSTAL STRUCTURE OF S-ADENOSYLMETHIONINE SYNTHETASE | | Descriptor: | COBALT (II) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
1XGS
 
 | |
1UVQ
 
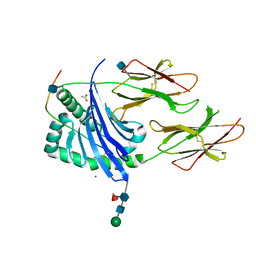 | | Crystal structure of HLA-DQ0602 in complex with a hypocretin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, GLYCINE, ... | | Authors: | Siebold, C, Hansen, B.E, Wyer, J.R, Harlos, K, Esnouf, R.E, Svejgaard, A, Bell, J.I, Strominger, J.L, Jones, E.Y, Fugger, L. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Hla-Dq0602 that Protects Against Type 1 Diabetes and Confers Strong Susceptibility to Narcolepsy
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1UWF
 
 | | 1.7 A resolution structure of the receptor binding domain of the FimH adhesin from uropathogenic E. coli | | Descriptor: | FIMH PROTEIN, GLYCEROL, butyl alpha-D-mannopyranoside | | Authors: | Bouckaert, J, Berglund, J, Genst, E.D, Cools, L, Hung, C.-S, Wuhrer, M, Zavialov, A, Langermann, S, Hultgren, S, Wyns, L, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-02-05 | | Release date: | 2005-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Receptor Binding Studies Disclose a Novel Class of High-Affinity Inhibitors of the Escherichia Coli Fimh Adhesin.
Mol.Microbiol., 55, 2005
|
|
1XVA
 
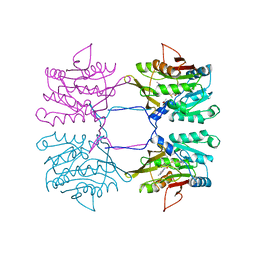 | | METHYLTRANSFERASE | | Descriptor: | ACETATE ION, GLYCINE N-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Fu, Z, Hu, Y, Konishi, K, Takata, Y, Ogawa, H, Gomi, T, Fujioka, M, Takusagawa, F. | | Deposit date: | 1996-07-20 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glycine N-methyltransferase from rat liver.
Biochemistry, 35, 1996
|
|
1UWM
 
 | | reduced ferredoxin 6 from Rhodobacter capsulatus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN VI | | Authors: | Sainz, G, Jakoncic, J, Sieker, L.C, Stojanoff, V, Sanishvili, N, Asso, M, Bertrand, P, Armengaud, J, Jouanneau, Y. | | Deposit date: | 2004-02-05 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a [2Fe-2S] Ferredoxin from Rhodobacter Capsulatus Likely Involved in Fe-S Cluster Biogenesis and Conformational Changes Observed Upon Reduction.
J.Biol.Inorg.Chem., 11, 2006
|
|
1UXA
 
 | | ADENOVIRUS AD37 FIBRE HEAD in complex with sialyl-lactose | | Descriptor: | ACETATE ION, FIBER PROTEIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
