1SOP
 
 | |
1SOQ
 
 | | Crystal structure of the transthyretin mutant A108Y/L110E solved in space group C2 | | Descriptor: | Transthyretin | | Authors: | Hornberg, A, Olofsson, A, Eneqvist, T, Lundgren, E, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-15 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The beta-strand D of transthyretin trapped in two discrete conformations
Biochim.Biophys.Acta, 1700, 2004
|
|
1SOR
 
 | | Aquaporin-0 membrane junctions reveal the structure of a closed water pore | | Descriptor: | Aquaporin-0 | | Authors: | Gonen, T, Sliz, P, Kistler, J, Cheng, Y, Walz, T. | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Aquaporin-0 membrane junctions reveal the structure of a closed water pore
Nature, 429, 2004
|
|
1SOS
 
 | | ATOMIC STRUCTURES OF WILD-TYPE AND THERMOSTABLE MUTANT RECOMBINANT HUMAN CU, ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE, ... | | Authors: | Parge, H.E, Hallewell, R.A, Tainer, J.A. | | Deposit date: | 1992-02-11 | | Release date: | 1993-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structures of wild-type and thermostable mutant recombinant human Cu,Zn superoxide dismutase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1SOT
 
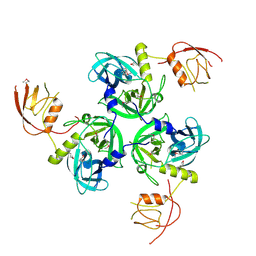 | | Crystal Structure of the DegS stress sensor | | Descriptor: | Protease degS | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
1SOU
 
 | | NMR structure of Aquifex aeolicus 5,10-methenyltetrahydrofolate synthetase: Northeast Structural Genomics Consortium Target QR46 | | Descriptor: | 5,10-methenyltetrahydrofolate synthetase | | Authors: | Cort, J.R, Chiang, Y, Acton, T, Wu, M, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Aquifex aeolicus 5,10-methenyltetrahydrofolate synthetase: Northeast Structural Genomics Consortium Target QR46
To be Published
|
|
1SOV
 
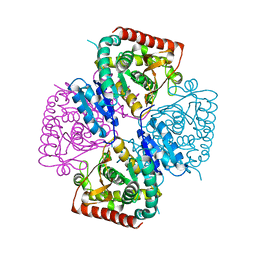 | |
1SOW
 
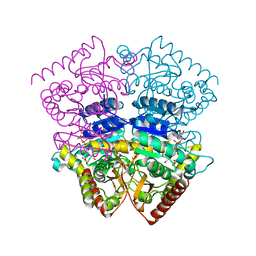 | |
1SOX
 
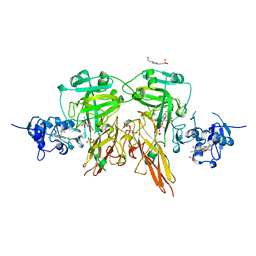 | | SULFITE OXIDASE FROM CHICKEN LIVER | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MOLYBDENUM ATOM, ... | | Authors: | Kisker, C, Schindelin, H, Rees, D.C. | | Deposit date: | 1997-12-31 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase.
Cell(Cambridge,Mass.), 91, 1997
|
|
1SOY
 
 | | Solution structure of the bacterial frataxin orthologue, CyaY | | Descriptor: | CyaY protein | | Authors: | Nair, M, Adinolfi, S, Pastore, C, Kelly, G, Temussi, P, Pastore, A. | | Deposit date: | 2004-03-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Bacterial Frataxin Ortholog, CyaY; Mapping the Iron Binding Sites
Structure, 12, 2004
|
|
1SOZ
 
 | | Crystal Structure of DegS protease in complex with an activating peptide | | Descriptor: | Protease degS, activating peptide | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-16 | | Release date: | 2004-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
1SP0
 
 | | Solution Structure of apoCox11 | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1SP1
 
 | |
1SP2
 
 | |
1SP3
 
 | | Crystal structure of octaheme cytochrome c from Shewanella oneidensis | | Descriptor: | HEME C, THIOCYANATE ION, cytochrome c, ... | | Authors: | Mowat, C.G, Rothery, E, Miles, C.S, McIver, L, Doherty, M.K, Drewette, K, Taylor, P, Walkinshaw, M.D, Chapman, S.K, Reid, G.A. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SP4
 
 | | Crystal structure of NS-134 in complex with bovine cathepsin B: a two headed epoxysuccinyl inhibitor extends along the whole active site cleft | | Descriptor: | Cathepsin B, methyl N-[(2S)-4-{[(1S)-1-{[(2S)-2-carboxypyrrolidin-1-yl]carbonyl}-3-methylbutyl]amino}-2-hydroxy-4-oxobutanoyl]-L-leucylglycylglycinate | | Authors: | Stern, I, Schaschke, N, Moroder, L, Turk, D. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of NS-134 in complex with bovine cathepsin B: a two-headed epoxysuccinyl inhibitor extends along the entire active-site cleft.
Biochem.J., 381, 2004
|
|
1SP5
 
 | | Crystal structure of HIV-1 protease complexed with a product of autoproteolysis | | Descriptor: | 5-mer peptide from Protease, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Vondrackova, E, Hasek, J, Jaskolski, M, Rezacova, P, Dohnalek, J, Skalova, T, Petrokova, H, Duskova, J, Brynda, J, Sedlacek, J. | | Deposit date: | 2004-03-16 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Product of enzymatic self-cleavage bound in the active site of HIV protease
To be Published
|
|
1SP6
 
 | | A DNA duplex containing a cholesterol adduct (alpha-anomer) | | Descriptor: | 5'-D(*CP*CP*AP*CP*(HOL)P*GP*GP*AP*AP*C)-3', 5'-D(GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3' | | Authors: | Gomez-Pinto, I, Cubero, E, Kalko, S.G, Monaco, V, van der Marel, G, van Boom, J.H, Orozco, M, Gonzalez, C. | | Deposit date: | 2004-03-16 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Effect of bulky lesions on DNA: Solution structure of a DNA duplex containing a cholesterol adduct.
J.Biol.Chem., 279, 2004
|
|
1SP7
 
 | | Structure of the Cys-rich C-terminal domain of Hydra minicollagen | | Descriptor: | mini-collagen | | Authors: | Meier, S, Haussinger, D, Pokidysheva, E, Bachinger, H.P, Grzesiek, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Determination of a high-precision NMR structure of the minicollagen cysteine rich domain from Hydra and characterization of its disulfide bond formation.
Febs Lett., 569, 2004
|
|
1SP8
 
 | | 4-Hydroxyphenylpyruvate Dioxygenase | | Descriptor: | 4-Hydroxyphenylpyruvate Dioxygenase, FE (II) ION | | Authors: | Fritze, I.M, Linden, L, Freigang, J, Auerbach, G, Huber, R, Steinbacher, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of Zea mays and Arabidopsis 4-Hydroxyphenylpyruvate Dioxygenase
Plant physiol., 134, 2004
|
|
1SP9
 
 | | 4-Hydroxyphenylpyruvate Dioxygenase | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, FE (II) ION | | Authors: | Fritze, I.M, Linden, L, Freigang, J, Auerbach, G, Huber, R, Steinbacher, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structures of Zea mays and Arabidopsis 4-Hydroxyphenylpyruvate Dioxygenase
Plant Physiol., 134, 2004
|
|
1SPA
 
 | | ROLE OF ASP222 IN THE CATALYTIC MECHANISM OF ESCHERICHIA COLI ASPARTATE AMINOTRANSFERASE: THE AMINO ACID RESIDUE WHICH ENHANCES THE FUNCTION OF THE ENZYME-BOUND COENZYME PYRIDOXAL 5'-PHOSPHATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, N-METHYL-4-DEOXY-4-AMINO-PYRIDOXAL-5-PHOSPHATE | | Authors: | Hinoue, Y, Yano, T, Metzler, D.E, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1993-01-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Asp222 in the catalytic mechanism of Escherichia coli aspartate aminotransferase: the amino acid residue which enhances the function of the enzyme-bound coenzyme pyridoxal 5'-phosphate.
Biochemistry, 31, 1992
|
|
1SPB
 
 | | SUBTILISIN BPN' PROSEGMENT (77 RESIDUES) COMPLEXED WITH A MUTANT SUBTILISIN BPN' (266 RESIDUES). CRYSTAL PH 4.6. CRYSTALLIZATION TEMPERATURE 20 C DIFFRACTION TEMPERATURE-160 C | | Descriptor: | SODIUM ION, SUBTILISIN BPN', SUBTILISIN BPN' PROSEGMENT | | Authors: | Gallagher, D.T, Gilliland, G.L, Wang, L, Bryan, P.N. | | Deposit date: | 1995-06-21 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The prosegment-subtilisin BPN' complex: crystal structure of a specific 'foldase'.
Structure, 3, 1995
|
|
1SPD
 
 | |
1SPE
 
 | | SPERM WHALE NATIVE CO MYOGLOBIN AT PH 4.0, TEMP 4C | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
