2D2S
 
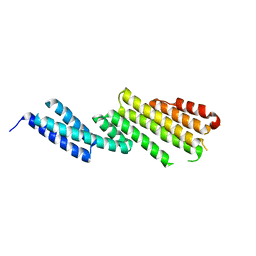 | | Crystal Structure of the Exo84p C-terminal Domains | | Descriptor: | Exocyst complex component EXO84 | | Authors: | Dong, G, Hutagalung, A.H, Fu, C, Novick, P, Reinisch, K.M. | | Deposit date: | 2005-09-16 | | Release date: | 2005-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif
Nat.Struct.Mol.Biol., 12, 2005
|
|
2F8D
 
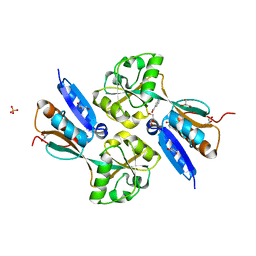 | | BenM effector-Binding domain crystallized from high pH conditions | | Descriptor: | BENZOIC ACID, GLYCEROL, HTH-type transcriptional regulator benM, ... | | Authors: | Ezezika, O.C, Haddad, S, Neidle, E.L, Momany, C. | | Deposit date: | 2005-12-02 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Oligomerization of BenM, a LysR-type transcriptional regulator: structural basis for the aggregation of proteins in this family.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2YCL
 
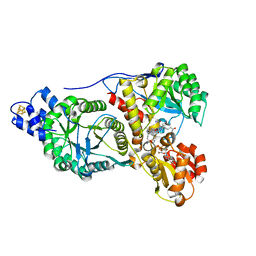 | | complete structure of the corrinoid,iron-sulfur protein including the N-terminal domain with a 4Fe-4S cluster | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE CORRINOID/IRON-SULFUR PROTEIN, GAMMA SUBUNIT, CO DEHYDROGENASE/ACETYL-COA SYNTHASE, ... | | Authors: | Goetzl, S, Jeoung, J.H, Hennig, S.E, Dobbek, H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Electron and Methyl-Group Transfer in a Methyltransferase System Operating in the Reductive Acetyl-Coa Pathway
J.Mol.Biol., 411, 2011
|
|
2YFV
 
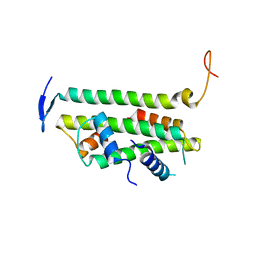 | |
3AHT
 
 | | Crystal structure of rice BGlu1 E176Q mutant in complex with laminaribiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Chuenchor, W, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Svasti, J, Ketudat Cairns, J.R. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3AMQ
 
 | | E134C-Cellobiose co-crystal of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AHV
 
 | | Semi-active E176Q mutant of rice bglu1 covalent complex with 2-deoxy-2-fluoroglucoside | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase 7, ... | | Authors: | Chuenchor, W, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Svasti, J, Ketudat Cairns, J.R. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3AMN
 
 | | E134C-Cellobiose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMH
 
 | | crystal structure of cellulase 12A from Thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3ASF
 
 | | MamA MSR-1 C2 | | Descriptor: | Magnetosome protein MamA, SULFATE ION | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2010-12-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AS8
 
 | | MamA MSR-1 P41212 | | Descriptor: | Magnetosome protein MamA, SULFATE ION | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2010-12-10 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AXE
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in complex with cellotetraose (cellobiose density was observed) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION, ... | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-04 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
3BAP
 
 | |
2ZL4
 
 | |
3BKH
 
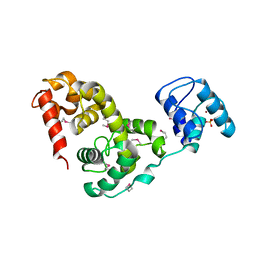 | | Crystal structure of the bacteriophage phiKZ lytic transglycosylase, gp144 | | Descriptor: | NICKEL (II) ION, SULFATE ION, lytic transglycosylase | | Authors: | Fokine, A, Miroshnikov, K.A, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the bacteriophage phi KZ lytic transglycosylase gp144.
J.Biol.Chem., 283, 2008
|
|
3C86
 
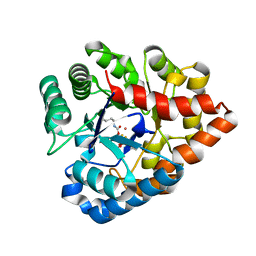 | | OpdA from agrobacterium radiobacter with bound product diethyl thiophosphate from crystal soaking with tetraethyl dithiopyrophosphate- 1.8 A | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, FE (II) ION, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2008-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
2ZD9
 
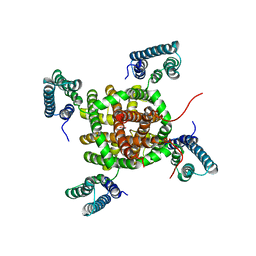 | |
2XQ1
 
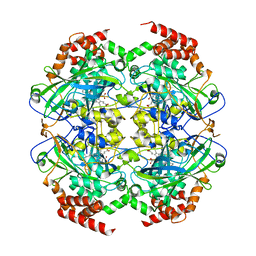 | | Crystal structure of peroxisomal catalase from the yeast Hansenula polymorpha | | Descriptor: | PEROXISOMAL CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Penya-Soler, E, Vega, M.C, Wilmanns, M, Williams, C.P. | | Deposit date: | 2010-08-31 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Features of Peroxisomal Catalase from the Yeast Hansenula Polymorpha
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XFL
 
 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Sun, H, Bricogne, G, Liang, Z, Lescar, J. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
2XEM
 
 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | (3E,5E,7E,9E,11E,13E)-pentadeca-3,5,7,9,11,13-hexaen-2-one, DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Bricogne, G, Liang, Z.X, Lescar, J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
3AI3
 
 | | The crystal structure of L-Sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose | | Descriptor: | L-sorbose, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-sorbose reductase, ... | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3ALX
 
 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (MV-H(L482R)-SLAM(N102H/R108Y) fusion) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin,LINKER,CDw150 | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-09 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
2ZEB
 
 | | Potent, Nonpeptide Inhibitors of Human Mast Cell Tryptase | | Descriptor: | 1-(1'-{[3-(methylsulfanyl)-2-benzothiophen-1-yl]carbonyl}spiro[1-benzofuran-3,4'-piperidin]-5-yl)methanamine, Tryptase beta 2 | | Authors: | Spurlino, J.C, Lewandowski, F, Milligan, C. | | Deposit date: | 2007-12-08 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3AI1
 
 | | The crystal structure of L-sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose reveals the structure bases of its catalytic mechanism and high substrate selectivity | | Descriptor: | NADPH-sorbose reductase | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3BEZ
 
 | |
