4Z90
 
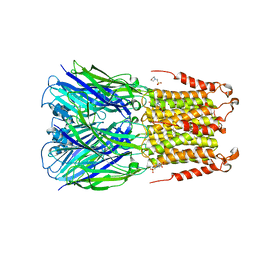 | | ELIC bound with the anesthetic isoflurane in the resting state | | Descriptor: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Chen, Q, Kinde, M, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
4Z0B
 
 | | Crystal Structure of the Fab Fragment of Anti-ofloxacin Antibody and Exploration Its Receptor Binding Site | | Descriptor: | PHOSPHATE ION, antibody heavy chain, antibody light chain | | Authors: | He, K, Du, X, Sheng, W, Zhou, X, Wang, J, Wang, S. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Fab Fragment of an Anti-ofloxacin Antibody and Exploration of Its Specific Binding.
J.Agric.Food Chem., 64, 2016
|
|
1MDZ
 
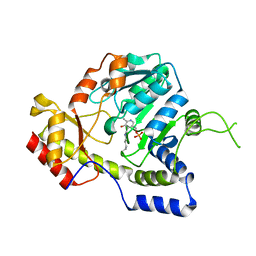 | | Crystal structure of ArnB aminotransferase with cycloserine and pyridoxal 5' phosphate | | Descriptor: | ArnB aminotransferase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
4Z9X
 
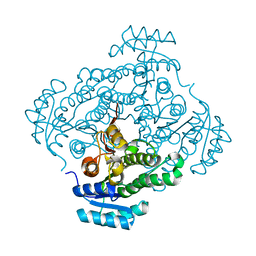 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus pyogenes | | Descriptor: | Gluconate 5-dehydrogenase | | Authors: | Maruyama, Y, Takase, R, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
1M9X
 
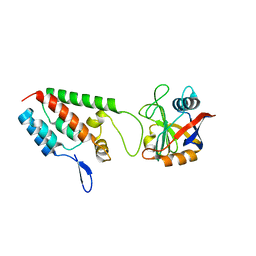 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A,A88M,G89A Complex. | | Descriptor: | Cyclophilin A, HIV-1 Capsid | | Authors: | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2002-07-30 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
4Z0J
 
 | | F96S/S73A Double mutant of Plasmodium Falciparum Triosephosphate Isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
1MEC
 
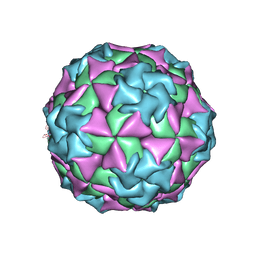 | |
4ZA4
 
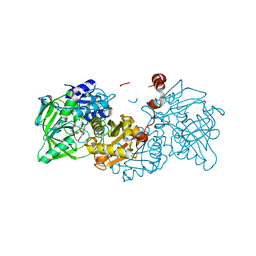 | | Structure of A. niger Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
1MHU
 
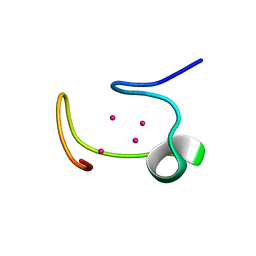 | | THE THREE-DIMENSIONAL STRUCTURE OF HUMAN [113CD7] METALLOTHIONEIN-2 IN SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Messerle, B.A, Schaeffer, A, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human [113Cd7]metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
1MF1
 
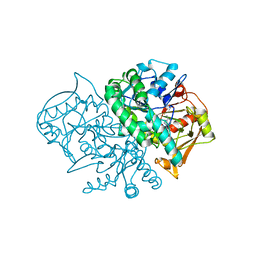 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
4Z0S
 
 | |
1MAL
 
 | |
4ZLF
 
 | | Cellobionic acid phosphorylase - cellobionic acid complex | | Descriptor: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4Z13
 
 | | Recombinantly expressed latent aurone synthase (polyphenol oxidase) co-crystallized with hexatungstotellurate(VI) and soaked in H2O2 | | Descriptor: | 6-tungstotellurate(VI), Aurone synthase, COPPER (II) ION, ... | | Authors: | Molitor, C, Mauracher, S.G, Rompel, A. | | Deposit date: | 2015-03-26 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Aurone synthase is a catechol oxidase with hydroxylase activity and provides insights into the mechanism of plant polyphenol oxidases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1MFG
 
 | |
4Z1I
 
 | | Crystal structure of human Trap1 with AMPPNP | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, MAGNESIUM ION, ... | | Authors: | Lee, C, Park, H.K, Ryu, J.H, Kang, B.H. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Development of a Mitochondria-Targeted Hsp90 Inhibitor Based on the Crystal Structures of Human TRAP1
J.Am.Chem.Soc., 137, 2015
|
|
1MBX
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ClpSN WITH TRANSITION METAL ION BOUND | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, CHLORIDE ION, ... | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of the AAA+ Chaperone, ClpA
J.Biol.Chem., 277, 2002
|
|
4ZLX
 
 | |
1MFL
 
 | |
4Z2F
 
 | | Sclerotium Rolfsii lectin variant (SSR2) with fine carbohydrate specificity | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, lectin variant 2 | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
1MCD
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-D-PHE-B-ALA-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
4Z2K
 
 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 32 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {7-[N'-(methylcarbamoyl)carbamimidamido]heptyl}carbamate, Chitinase B, GLYCEROL | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
1MFT
 
 | | Crystal Structure Of Four-Helix Bundle Model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Lahr, S.J, Stayrook, S.E, North, B, Kaplan, J, Geremia, S, DeGrado, W. | | Deposit date: | 2002-08-13 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
4ZM3
 
 | | Crystal structure of PLP-Dependent 3-Aminobenzoate Synthase PctV wild-type | | Descriptor: | Aminotransferase, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Hirayama, A, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2015-05-02 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Mechanism-Based Trapping of the Quinonoid Intermediate by Using the K276R Mutant of PLP-Dependent 3-Aminobenzoate Synthase PctV in the Biosynthesis of Pactamycin.
Chembiochem, 16, 2015
|
|
1MD0
 
 | |
