1IFD
 
 | |
1IFJ
 
 | |
1IFI
 
 | |
1OLA
 
 | |
1PAR
 
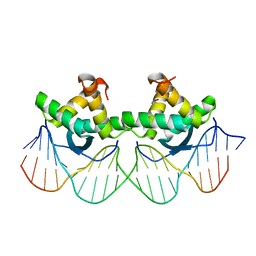 | | DNA RECOGNITION BY BETA-SHEETS IN THE ARC REPRESSOR-OPERATOR CRYSTAL STRUCTURE | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*T P*AP*CP*TP*AP*T)- 3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*T P*AP*TP*CP*AP*T)- 3'), PROTEIN (ARC REPRESSOR) | | Authors: | Raumann, B.E, Rould, M.A, Pabo, C.O, Sauer, R.T. | | Deposit date: | 1994-03-22 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA recognition by beta-sheets in the Arc repressor-operator crystal structure.
Nature, 367, 1994
|
|
1AST
 
 | |
2DHC
 
 | |
1BYA
 
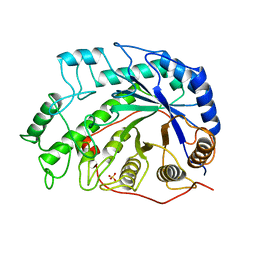 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1PMY
 
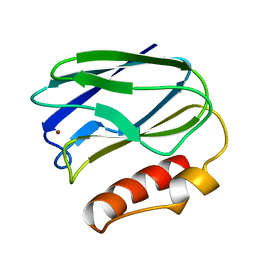 | | REFINED CRYSTAL STRUCTURE OF PSEUDOAZURIN FROM METHYLOBACTERIUM EXTORQUENS AM1 AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Kai, Y, Harada, S, Kasai, N, Ohshiro, Y, Suzuki, S, Kohzuma, T, Tobari, J. | | Deposit date: | 1994-01-28 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Refined crystal structure of pseudoazurin from Methylobacterium extorquens AM1 at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1FLP
 
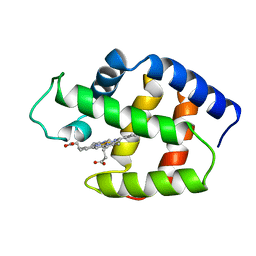 | | STRUCTURE OF THE SULFIDE-REACTIVE HEMOGLOBIN FROM THE CLAM LUCINA PECTINATA: CRYSTALLOGRAPHIC ANALYSIS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | HEMOGLOBIN I (AQUO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rizzi, M, Wittenberg, J.B, Ascenzi, P, Fasano, M, Coda, A, Bolognesi, M. | | Deposit date: | 1994-05-16 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the sulfide-reactive hemoglobin from the clam Lucina pectinata. Crystallographic analysis at 1.5 A resolution.
J.Mol.Biol., 244, 1994
|
|
1HMA
 
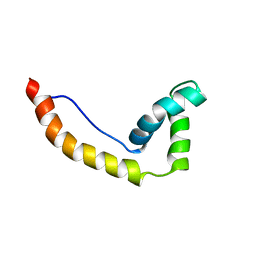 | | THE SOLUTION STRUCTURE AND DYNAMICS OF THE DNA BINDING DOMAIN OF HMG-D FROM DROSOPHILA MELANOGASTER | | Descriptor: | HMG-D | | Authors: | Jones, D.N.M, Searles, M.A, Shaw, G.L, Churchill, M.E.A, Ner, S.S, Keeler, J, Travers, A.A, Neuhaus, D. | | Deposit date: | 1994-05-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster.
Structure, 2, 1994
|
|
1SYB
 
 | | TRANSFER OF A BETA-TURN STRUCTURE TO A NEW PROTEIN CONTEXT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Hynes, T.R, Kautz, R.A, Goodman, M.A, Gill, J.F, Fox, R.O. | | Deposit date: | 1994-01-07 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transfer of a beta-turn structure to a new protein context.
Nature, 339, 1989
|
|
1TRI
 
 | |
1HTF
 
 | |
1BYB
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BVH
 
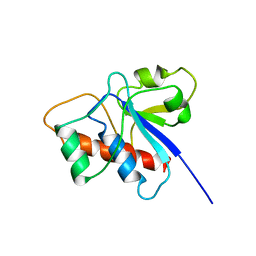 | | SOLUTION STRUCTURE OF A LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE | | Descriptor: | ACID PHOSPHATASE | | Authors: | Logan, T.M, Zhou, M.-M, Nettesheim, D.G, Meadows, R.P, Van Etten, R.L, Fesik, S.W. | | Deposit date: | 1994-05-03 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a low molecular weight protein tyrosine phosphatase.
Biochemistry, 33, 1994
|
|
2BMH
 
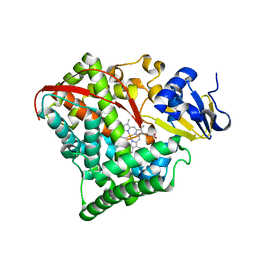 | |
1BMD
 
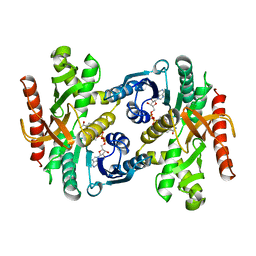 | |
1AKA
 
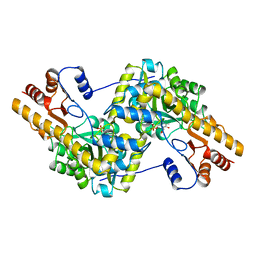 | |
2CHT
 
 | |
1BPE
 
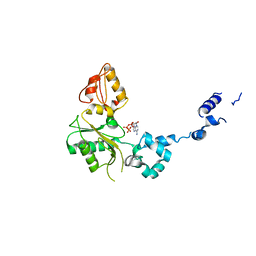 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA; EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA POLYMERASE BETA | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-04-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
1BYC
 
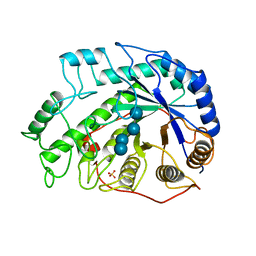 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BYD
 
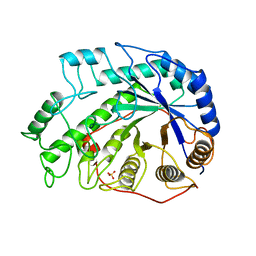 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1ACY
 
 | |
1BRB
 
 | |
