2BWS
 
 | | His243Ala Escherichia coli Aminopeptidase P | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, XAA-PRO AMINOPEPTIDASE P | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2C0R
 
 | |
1NKH
 
 | | Crystal structure of Lactose synthase complex with UDP and Manganese | | Descriptor: | ALPHA-LACTALBUMIN, BETA-1,4-GALACTOSYLTRANSFERASE, CALCIUM ION, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2003-01-03 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lactose synthase reveals a large conformational
change in its catalytic component, the beta-1,4-galactosyltransferase
J.Mol.Biol., 310, 2001
|
|
1IRF
 
 | | INTERFERON REGULATORY FACTOR-2 DNA BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERFERON REGULATORY FACTOR-2 | | Authors: | Furui, J, Uegaki, K, Yamazaki, T, Shirakawa, M, Swindells, M.B, Harada, H, Taniguchi, T, Kyogoku, Y. | | Deposit date: | 1997-11-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IRF-2 DNA-binding domain: a novel subgroup of the winged helix-turn-helix family.
Structure, 6, 1998
|
|
2C02
 
 | | Crystal Structures of Eosinophil-derived Neurotoxin in Complex with the Inhibitors 5'-ATP, Ap3A, Ap4A and Ap5A | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, NONSECRETORY RIBONUCLEASE | | Authors: | Baker, M.D, Holloway, D.E, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Eosinophil-Derived Neurotoxin (Edn) in Complex with the Inhibitors 5'- ATP, Ap(3)A, Ap(4)A, and Ap(5)A.
Biochemistry, 45, 2006
|
|
1N7Y
 
 | | STREPTAVIDIN MUTANT N23E AT 1.96A | | Descriptor: | Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-11-18 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NKW
 
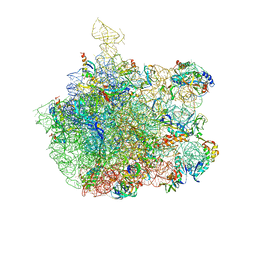 | | Crystal Structure Of The Large Ribosomal Subunit From Deinococcus Radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Harms, J.M, Schluenzen, F, Zarivach, R, Bashan, A, Gat, S, Agmon, I, Bartels, H, Franceschi, F, Yonath, A. | | Deposit date: | 2003-01-05 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | High resolution structure of the large ribosomal subunit from a mesophilic eubacterium
Cell(Cambridge,Mass.), 107, 2001
|
|
1N86
 
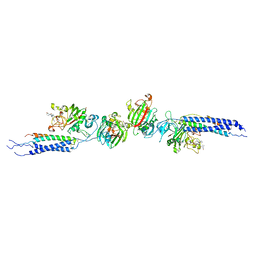 | | Crystal structure of human D-dimer from cross-linked fibrin complexed with GPR and GHRPLDK peptide ligands. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, Fibrin alpha/alpha-E chain, ... | | Authors: | Yang, Z, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-11-19 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of fragment double-D from cross-linked lamprey fibrin reveals isopeptide linkages across an unexpected D-D interface.
Biochemistry, 41, 2002
|
|
2C23
 
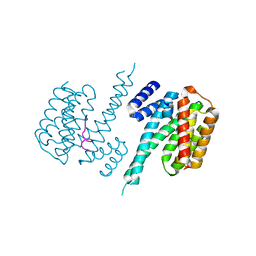 | | 14-3-3 Protein Beta (Human) in complex with exoenzyme S peptide | | Descriptor: | 14-3-3 BETA/ALPHA, EXOENZYME S PEPTIDE | | Authors: | Elkins, J.M, Schoch, G.A, Yang, X, Sundstrom, M, Arrowsmith, C, Edwards, A, Doyle, D.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1IS7
 
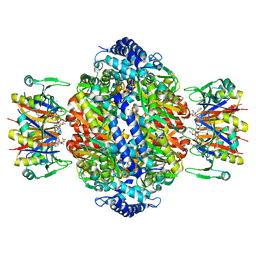 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1NFU
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR132747 | | Descriptor: | 3-({4-[(6-CHLORO-1-BENZOTHIEN-2-YL)SULFONYL]-2-OXOPIPERAZIN-1-YL}METHYL)BENZENECARBOXIMIDAMIDE, CALCIUM ION, COAGULATION FACTOR XA, ... | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-12-16 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular structures of human Factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket.
J.Med.Chem., 46, 2003
|
|
1NFY
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR200095 | | Descriptor: | 4-({4-[(6-CHLORO-1-BENZOTHIEN-2-YL)SULFONYL]-2-OXOPIPERAZIN-1-YL}METHYL)BENZENECARBOXIMIDAMIDE, CALCIUM ION, Coagulation factor XA, ... | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-12-16 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular structures of human Factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket.
J.Med.Chem., 46, 2003
|
|
1ILE
 
 | | ISOLEUCYL-TRNA SYNTHETASE | | Descriptor: | ISOLEUCYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Nureki, O, Vassylyev, D.G, Tateno, M, Shimada, A, Nakama, T, Fukai, S, Konno, M, Schimmel, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-02-24 | | Release date: | 1999-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme structure with two catalytic sites for double-sieve selection of substrate.
Science, 280, 1998
|
|
1NH3
 
 | | Human Topoisomerase I Ara-C Complex | | Descriptor: | 5'-D(*(GNG)P*GP*AP*AP*AP*AP*AP*UP*UP*UP*UP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*UP*(UBB))-3', 5'-D(*AP*AP*AP*AP*AP*TP*UP*UP*UP*UP*CP*(CAR)P*AP*AP*GP*UP*CP*UP*UP*UP*UP*T)-3', ... | | Authors: | Chrencik, J.E, Burgin, A.B, Pommier, Y, Stewart, L, Redinbo, M.R. | | Deposit date: | 2002-12-18 | | Release date: | 2003-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Impact of the Leukemia Drug 1-beta-D-Arabinofuranosylcytosine (Ara-C) on the Covalent Human Topoisomerase I-DNA Complex
J.Biol.Chem., 278, 2003
|
|
1NBI
 
 | |
2BWW
 
 | | His350Ala Escherichia coli Aminopeptidase P | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, AMINOPEPTIDASE P, CITRATE ANION, ... | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
1IMD
 
 | |
1IX8
 
 | | Aspartate Aminotransferase Active Site Mutant V39F/N194A | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
2C3E
 
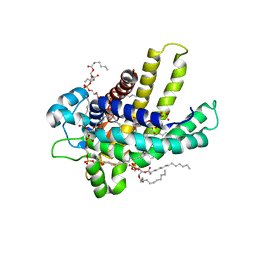 | | The bovine mitochondrial ADP-ATP carrier | | Descriptor: | ADP/ATP TRANSLOCASE 1, CARDIOLIPIN, Carboxyatractyloside | | Authors: | Nury, H, Dahout-Gonzalez, C, Trezeguet, V, Lauquin, G, Brandolin, G, Pebay-Peyroula, E. | | Deposit date: | 2005-10-06 | | Release date: | 2005-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Lipid-Mediated Interactions between Mitochondrial Adp/ATP Carrier Monomers.
FEBS Lett., 579, 2005
|
|
1NCC
 
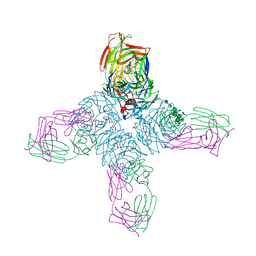 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
1ND6
 
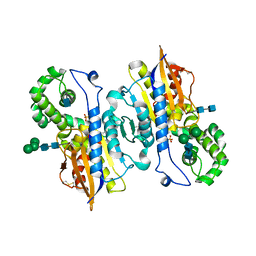 | | Crystal Structures of Human Prostatic Acid Phosphatase in Complex with a Phosphate Ion and alpha-Benzylaminobenzylphosphonic Acid Update the Mechanistic Picture and Offer New Insights into Inhibitor Design | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Ortlund, E, LaCount, M.W, Lebioda, L. | | Deposit date: | 2002-12-07 | | Release date: | 2002-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of human prostatic acid phosphatase in complex with a
phosphate ion and alpha-benzylaminobenzylphosphonic acid update the
mechanistic picture and offer new insights into inhibitor design
Biochemistry, 42, 2003
|
|
2BPO
 
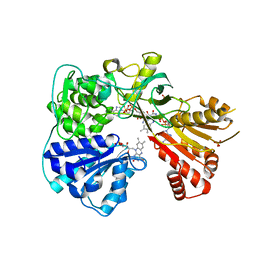 | | Crystal structure of the yeast CPR triple mutant: D74G, Y75F, K78A. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yermalitskaya, L.V, Kim, Y, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-04-21 | | Release date: | 2006-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Yeast Cpr Triple Mutant: D74G, Y75F, K78A.
To be Published
|
|
2C5R
 
 | | The structure of phage phi29 replication organizer protein p16.7 in complex with double stranded DNA | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*GP*GP*AP)-3', 5'-D(*TP*CP*CP*AP*CP*CP*GP*GP)-3', EARLY PROTEIN P16.7 | | Authors: | Albert, A, Jimenez, M, Munoz-Espin, D, Asensio, J.L, Hermoso, J.A, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Membrane Anchorage of Viral Phi 29 DNA During Replication.
J.Biol.Chem., 280, 2005
|
|
1HEC
 
 | |
2BFN
 
 | | The crystal structure of the complex of the haloalkane dehalogenase LinB with the product of dehalogenation reaction 1,2-dichloropropane. | | Descriptor: | (2S)-2,3-DICHLOROPROPAN-1-OL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Banas, P, Otyepka, M, Jerabek, P, Vevodova, J, Bohac, M, Damborsky, J. | | Deposit date: | 2004-12-09 | | Release date: | 2006-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Weak Activity of Haloalkane Dehalogenase Linb with 1,2,3-Trichloropropane Revealed by X-Ray Crystallography and Microcalorimetry
Appl.Environ.Microbiol., 73, 2007
|
|
