1HUT
 
 | | THE STRUCTURE OF ALPHA-THROMBIN INHIBITED BY A 15-MER SINGLE-STRANDED DNA APTAMER | | Descriptor: | ALPHA-Thrombin heavy chain, ALPHA-Thrombin light chain, D-phenylalanyl-N-[(3S)-6-carbamimidamido-1-chloro-2-oxohexan-3-yl]-L-prolinamide, ... | | Authors: | Padmanabhan, K, Padmanabhan, K.P, Ferrara, J.D, Sadler, J.E, Tulinsky, A. | | Deposit date: | 1993-05-27 | | Release date: | 1994-06-22 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer.
J.Biol.Chem., 268, 1993
|
|
1XIF
 
 | |
1XID
 
 | |
1STA
 
 | | ACCOMMODATION OF INSERTION MUTATIONS ON THE SURFACE AND IN THE INTERIOR OF STAPHYLOCOCCAL NUCLEASE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Keefe, L.J, Quirk, S, Gittis, A, Lattman, E.E. | | Deposit date: | 1994-01-17 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of insertion mutations on the surface and in the interior of staphylococcal nuclease.
Protein Sci., 3, 1994
|
|
1GLM
 
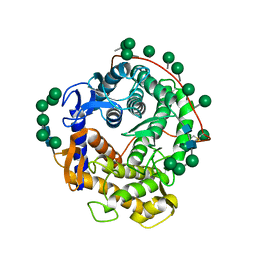 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | Descriptor: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-04-25 | | Release date: | 1994-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
1XIE
 
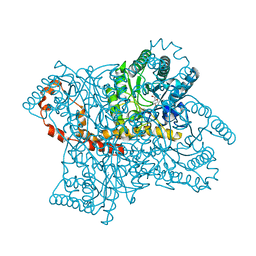 | |
1GLP
 
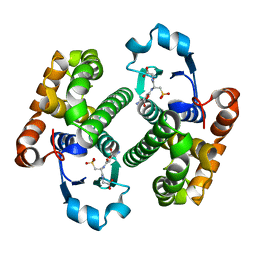 | |
1PHP
 
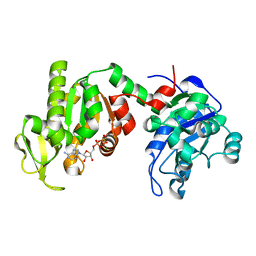 | |
1CPN
 
 | |
1XII
 
 | |
1XIJ
 
 | |
1PLK
 
 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PLJ
 
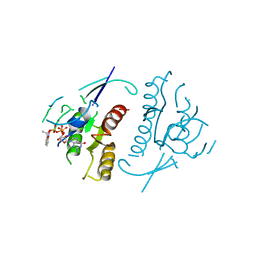 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE 5'-TRIPHOSPHATE P3-[1-(2-NITROPHENYL)ETHYL ESTER], MAGNESIUM ION | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PLL
 
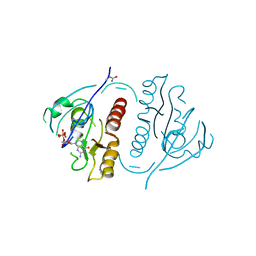 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1AST
 
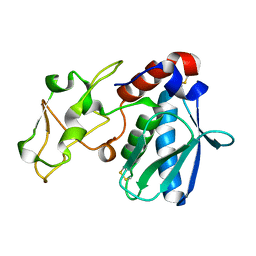 | |
1TRI
 
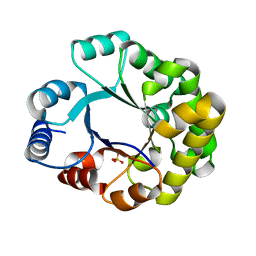 | |
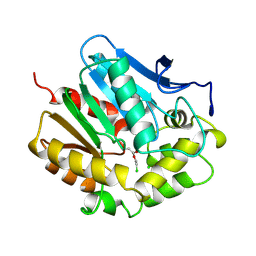
2DHD
 
 | |
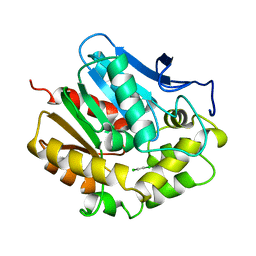
2DHC
 
 | |
1BYA
 
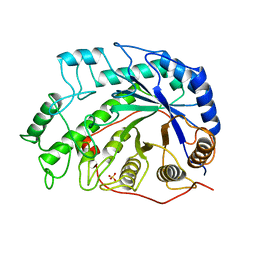 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BVH
 
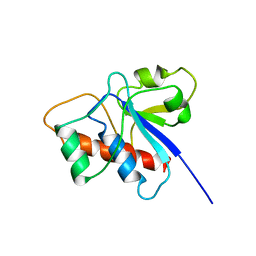 | | SOLUTION STRUCTURE OF A LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE | | Descriptor: | ACID PHOSPHATASE | | Authors: | Logan, T.M, Zhou, M.-M, Nettesheim, D.G, Meadows, R.P, Van Etten, R.L, Fesik, S.W. | | Deposit date: | 1994-05-03 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a low molecular weight protein tyrosine phosphatase.
Biochemistry, 33, 1994
|
|
1BYB
 
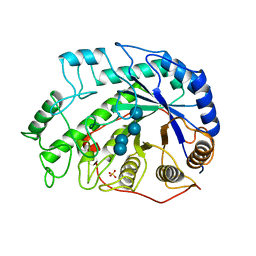 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
216L
 
 | |
1TYT
 
 | |
1CRQ
 
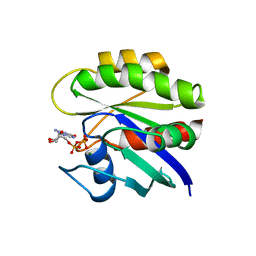 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | Deposit date: | 1993-11-24 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CHN
 
 | |
