9ICW
 
 | | DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; NATIVE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*CP*TP*GP*T)-3'), PROTEIN (DNA POLYMERASE BETA (E.C.2.7.7.7)), ... | | Authors: | Pelletier, H, Sawaya, M.R. | | Deposit date: | 1995-12-16 | | Release date: | 1996-11-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity
Biochemistry, 35, 1996
|
|
2MKB
 
 | |
1AB7
 
 | |
2LZQ
 
 | |
3NSN
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with TMG-chitotriomycin | | Descriptor: | 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
5YN8
 
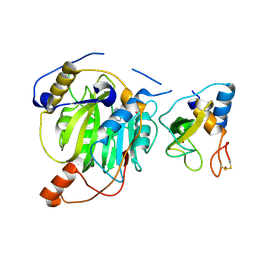 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNO
 
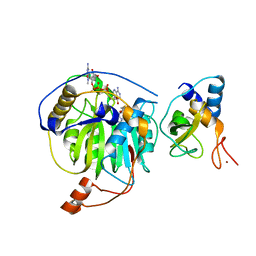 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
1RMV
 
 | | RIBGRASS MOSAIC VIRUS, FIBER DIFFRACTION | | Descriptor: | RIBGRASS MOSAIC VIRUS COAT PROTEIN, RIBGRASS MOSAIC VIRUS RNA | | Authors: | Wang, H, Stubbs, G. | | Deposit date: | 1997-02-11 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | FIBER DIFFRACTION (2.9 Å) | | Cite: | Molecular dynamics in refinement against fiber diffraction data.
Acta Crystallogr.,Sect.A, 49, 1993
|
|
5YN6
 
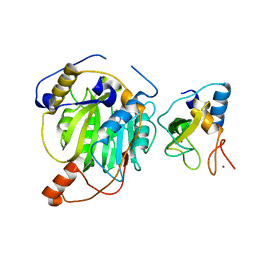 | | Crystal structure of MERS-CoV nsp10/nsp16 complex bound to SAM | | Descriptor: | S-ADENOSYLMETHIONINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
2W7J
 
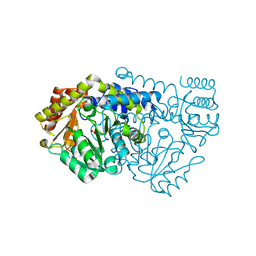 | | Crystal structure of Y61AbsSHMT Glycine external aldimine | | Descriptor: | GLYCINE, PYRIDOXAL-5'-PHOSPHATE, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Rajaram, V, Bhavani, B.S, Bisht, S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-12-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Importance of Tyrosine Residues of Bacillus Stearothermophilus Serine Hydroxymethyltransferase in Cofactor Binding and L-Allo-Thr Cleavage.
FEBS J., 275, 2008
|
|
2W7D
 
 | | Crystal structure of Y51FbsSHMT internal aldimine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Rajaram, V, Bhavani, B.S, Bisht, S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-12-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Importance of Tyrosine Residues of Bacillus Stearothermophilus Serine Hydroxymethyltransferase in Cofactor Binding and L-Allo-Thr Cleavage.
FEBS J., 275, 2008
|
|
5YN5
 
 | | Crystal structure of MERS-CoV nsp10/nsp16 complex | | Descriptor: | ZINC ION, nsp10 protein, nsp16 protein | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNF
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNP
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to sinefungin and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
2W7L
 
 | | Crystal structure of Y61AbsSHMT L-allo-Threonine external aldimine | | Descriptor: | ALLO-THREONINE, PYRIDOXAL-5'-PHOSPHATE, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Rajaram, V, Bhavani, B.S, Bisht, S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-12-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Importance of Tyrosine Residues of Bacillus Stearothermophilus Serine Hydroxymethyltransferase in Cofactor Binding and L-Allo-Thr Cleavage.
FEBS J., 275, 2008
|
|
2W7K
 
 | | Crystal structure of Y61AbsSHMT L-Serine external aldimine | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Rajaram, V, Bhavani, B.S, Bisht, S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-12-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Importance of Tyrosine Residues of Bacillus Stearothermophilus Serine Hydroxymethyltransferase in Cofactor Binding and L-Allo-Thr Cleavage.
FEBS J., 275, 2008
|
|
5YNB
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to Sinefungin | | Descriptor: | SINEFUNGIN, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNM
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
3J2V
 
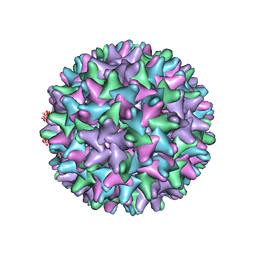 | | CryoEM structure of HBV core | | Descriptor: | PreC/core protein | | Authors: | Yu, X, Jin, L, Jih, J, Shih, C, Zhou, Z.H. | | Deposit date: | 2013-01-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 3.5 angstrom cryoEM Structure of Hepatitis B Virus Core Assembled from Full-Length Core Protein.
Plos One, 8, 2013
|
|
2W7F
 
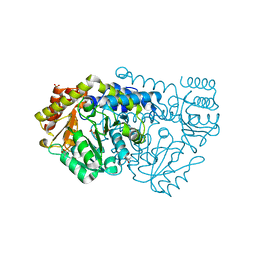 | | Crystal structure of Y51FbsSHMT L-Ser external aldimine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Rajaram, V, Bhavani, B.S, Bisht, S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-12-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Importance of Tyrosine Residues of Bacillus Stearothermophilus Serine Hydroxymethyltransferase in Cofactor Binding and L-Allo-Thr Cleavage.
FEBS J., 275, 2008
|
|
1R38
 
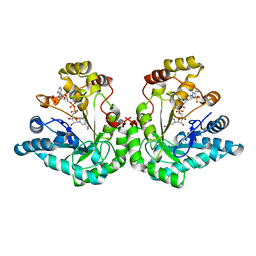 | | Crystal structure of H114A mutant of Candida tenuis xylose reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, xylose reductase | | Authors: | Kratzer, R, Kavanagh, K.L, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2003-09-30 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies of the enzymic mechanism of Candida tenuis xylose reductase (AKR 2B5): X-ray structure and catalytic reaction profile for the H113A mutant
Biochemistry, 43, 2004
|
|
7UJB
 
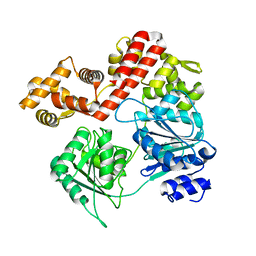 | | N-terminal domain deletion variant of Eta | | Descriptor: | DEAD/DEAH box RNA helicase | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2022-03-30 | | Release date: | 2022-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.12 Å) | | Cite: | The structure and activities of the archaeal transcription termination factor Eta detail vulnerabilities of the transcription elongation complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5O7D
 
 | | The X-ray structure of human R38M phosphoglycerate kinase 1 mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoglycerate kinase 1 | | Authors: | Ilari, A, Fiorillo, A, Cipollone, A, Petrosino, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
2W7I
 
 | | Crystal structure of Y61AbsSHMT internal aldimine | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Rajaram, V, Bhavani, B.S, Bisht, S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-12-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Importance of Tyrosine Residues of Bacillus Stearothermophilus Serine Hydroxymethyltransferase in Cofactor Binding and L-Allo-Thr Cleavage.
FEBS J., 275, 2008
|
|
7TL3
 
 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF431 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
