1MZ8
 
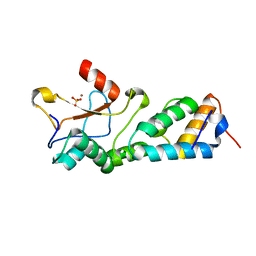 | | CRYSTAL STRUCTURES OF THE NUCLEASE DOMAIN OF COLE7/IM7 IN COMPLEX WITH A PHOSPHATE ION AND A ZINC ION | | Descriptor: | Colicin E7, Colicin E7 immunity protein, PHOSPHATE ION, ... | | Authors: | Sui, M.J, Tsai, L.C, Hsia, K.C, Doudeva, L.G, Ku, W.Y, Han, G.W, Yuan, H.S. | | Deposit date: | 2002-10-07 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal ions and phosphate binding in the H-N-H motif: crystal structures of the nuclease domain of ColE7/Im7 in complex with a phosphate ion and different divalent metal ions
PROTEIN SCI., 11, 2002
|
|
1MZM
 
 | |
1NBY
 
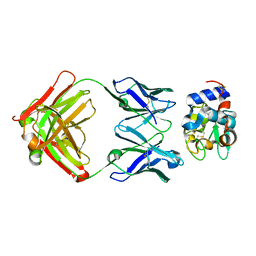 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K96A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
1NDA
 
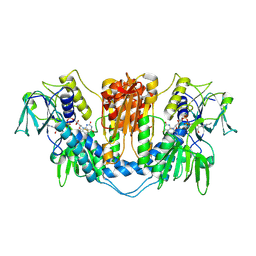 | | THE STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN THE OXIDIZED AND NADPH REDUCED STATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE OXIDOREDUCTASE | | Authors: | Lantwin, C.B, Kabsch, W, Pai, E.F, Schlichting, I, Krauth-Siegel, R.L. | | Deposit date: | 1993-07-02 | | Release date: | 1994-09-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of Trypanosoma cruzi trypanothione reductase in the oxidized and NADPH reduced state.
Proteins, 18, 1994
|
|
1NF1
 
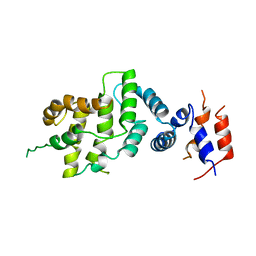 | | THE GAP RELATED DOMAIN OF NEUROFIBROMIN | | Descriptor: | PROTEIN (NEUROFIBROMIN) | | Authors: | Scheffzek, K, Ahmadian, M.R, Wiesmueller, L, Kabsch, W, Stege, P, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1998-07-08 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the GAP-related domain from neurofibromin and its implications.
EMBO J., 17, 1998
|
|
1N2C
 
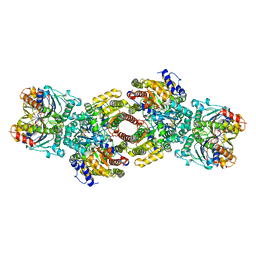 | | NITROGENASE COMPLEX FROM AZOTOBACTER VINELANDII STABILIZED BY ADP-TETRAFLUOROALUMINATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Schindelin, H, Kisker, C, Rees, D.C. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction.
Nature, 387, 1997
|
|
1NJX
 
 | |
1NK6
 
 | |
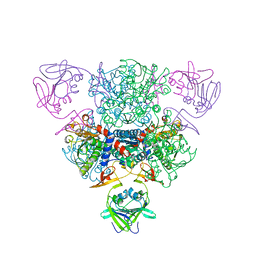
1NBE
 
 | |
1N7E
 
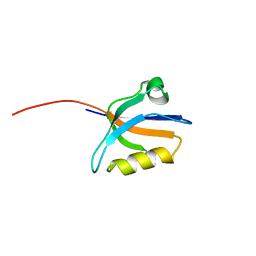 | | Crystal structure of the sixth PDZ domain of GRIP1 | | Descriptor: | AMPA receptor interacting protein GRIP | | Authors: | Im, Y.J, Park, S.H, Rho, S.H, Lee, J.H, Kang, G.B, Sheng, M, Kim, E, Eom, S.H. | | Deposit date: | 2002-11-14 | | Release date: | 2003-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization
J.BIOL.CHEM., 278, 2003
|
|
1N7Z
 
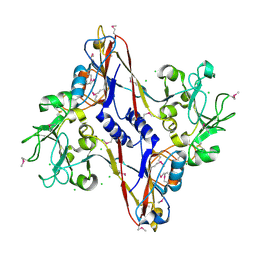 | | Structure and location of gene product 8 in the bacteriophage T4 baseplate | | Descriptor: | CHLORIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-18 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1NQI
 
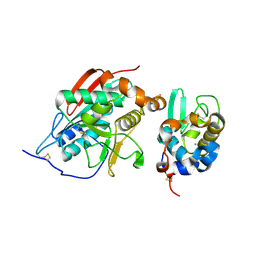 | | crystal structure of lactose synthase, a 1:1 complex between beta1,4-galactosyltransferase and alpha-lactalbumin in the presence of GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-LACTALBUMIN, BETA-1,4-GALACTOSYLTRANSFERASE, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2003-01-21 | | Release date: | 2003-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lactose synthase reveals a large conformational
change in its catalytic component, the beta-1,4-galactosyltransferase
J.Mol.Biol., 310, 2001
|
|
1O1K
 
 | |
1F30
 
 | | THE STRUCTURAL BASIS FOR DNA PROTECTION BY E. COLI DPS PROTEIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, ZINC ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2000-05-31 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Structural Basis for DNA Protection by E. coli Dps Protein
To be Published
|
|
1O4D
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78262. | | Descriptor: | 2-FORMYLPHENYL DIHYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O6O
 
 | |
1O86
 
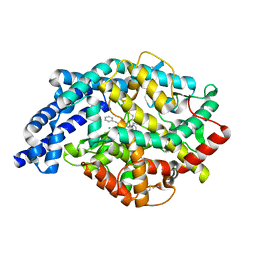 | | Crystal Structure of Human Angiotensin Converting Enzyme in complex with lisinopril. | | Descriptor: | ANGIOTENSIN CONVERTING ENZYME, CHLORIDE ION, GLYCINE, ... | | Authors: | Natesh, R, Schwager, S.L.U, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2002-11-25 | | Release date: | 2003-02-07 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Angiotensin-Converting Enzyme-Lisinopril Complex
Nature, 421, 2003
|
|
1F1H
 
 | |
1NOL
 
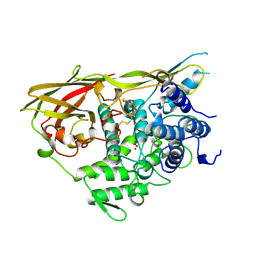 | | OXYGENATED HEMOCYANIN (SUBUNIT TYPE II) | | Descriptor: | CALCIUM ION, COPPER (II) ION, HEMOCYANIN (SUBUNIT TYPE II), ... | | Authors: | Hazes, B, Hol, W.G.J. | | Deposit date: | 1995-10-17 | | Release date: | 1996-03-08 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of deoxygenated Limulus polyphemus subunit II hemocyanin at 2.18 A resolution: clues for a mechanism for allosteric regulation.
Protein Sci., 2, 1993
|
|
1NOZ
 
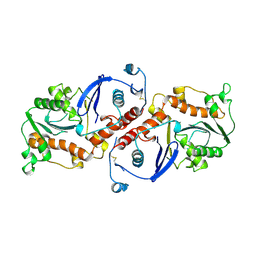 | | T4 DNA POLYMERASE FRAGMENT (RESIDUES 1-388) AT 110K | | Descriptor: | DNA POLYMERASE | | Authors: | Wang, J, Yu, P, Lin, T.C, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1996-02-16 | | Release date: | 1996-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions.
Biochemistry, 35, 1996
|
|
1NOD
 
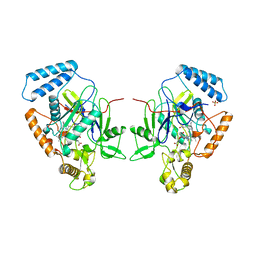 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH TETRAHYDROBIOPTERIN AND SUBSTRATE L-ARGININE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1998-03-05 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of nitric oxide synthase oxygenase dimer with pterin and substrate.
Science, 279, 1998
|
|
1NRI
 
 | | Crystal Structure of Putative Phosphosugar Isomerase HI0754 from Haemophilus influenzae | | Descriptor: | Hypothetical protein HI0754 | | Authors: | Kim, Y, Quartey, P, Ng, R, Zarembinski, T.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-24 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Hypothetical protein HI0754 from Haemophilus influenzae
To be Published
|
|
1NW7
 
 | | Structure of the beta class N6-adenine DNA methyltransferase RsrI bound to S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | CHLORIDE ION, MODIFICATION METHYLASE RSRI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
1NWQ
 
 | | CRYSTAL STRUCTURE OF C/EBPALPHA-DNA COMPLEX | | Descriptor: | 5'-D(*AP*AP*AP*CP*TP*GP*GP*AP*TP*TP*GP*CP*GP*CP*AP*AP*TP*AP*GP*GP*A)-3', 5'-D(*TP*TP*CP*CP*TP*AP*TP*TP*GP*CP*GP*CP*AP*AP*TP*CP*CP*AP*GP*TP*T)-3', CCAAT/enhancer binding protein alpha | | Authors: | Miller, M, Shuman, J.D, Sebastian, T, Dauter, Z, Johnson, P.F. | | Deposit date: | 2003-02-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for DNA Recognition by the Basic Region Leucine Zipper
Transcription Factor CCAAT/enhancer Binding Protein Alpha
J.Biol.Chem., 278, 2003
|
|
1NWG
 
 | | BETA-1,4-GALACTOSYLTRANSFERASE COMPLEX WITH ALPHA-LACTALBUMIN AND N-BUTANOYL-GLUCOAMINE | | Descriptor: | 2-(butanoylamino)-2-deoxy-beta-D-glucopyranose, Alpha-lactalbumin, CALCIUM ION, ... | | Authors: | Ramakrishnan, B, Shah, P.S, Qasba, P.K. | | Deposit date: | 2003-02-06 | | Release date: | 2003-02-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | ALPHA-LACTALBUMIN (LA) STIMULATES MILK
BETA-1,4-GALACTOSYLTRANSFERASE I (BETA 4GAL-T1) TO
TRANSFER GLUCOSE FROM UDP-GLUCOSE TO
N-ACETYLGLUCOSAMINE. CRYSTAL STRUCTURE OF BETA
4GAL-T1 X LA COMPLEX WITH UDP-GLC.
J.Biol.Chem., 276, 2001
|
|
