4FHV
 
 | |
7K87
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease in complex with SJ000986436 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2020-09-25 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
4FH5
 
 | |
4FHY
 
 | |
4FHP
 
 | | Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity - CaUTP bound | | Descriptor: | CALCIUM ION, GLYCEROL, Poly(A) RNA polymerase protein cid1, ... | | Authors: | Lunde, B.M, Magler, I, Meinhart, A. | | Deposit date: | 2012-06-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity.
Nucleic Acids Res., 40, 2012
|
|
4FHW
 
 | |
8G1U
 
 | | Structure of the methylosome-Lsm10/11 complex | | Descriptor: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | Authors: | Lin, M, Paige, A, Tong, L. | | Deposit date: | 2023-02-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
5VZB
 
 | | Post-catalytic complex of human Polymerase Mu (G433S) mutant with incoming UTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5VZH
 
 | | Post-catalytic complex of human Polymerase Mu (W434H) mutant with incoming UTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
3G86
 
 | | Hepatitis C virus polymerase NS5B (BK 1-570) with thiazine inhibitor | | Descriptor: | N-{3-[6-fluoro-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxido-4H-1,4-benzothiazin-7-yl}methanesulfonamide, NICKEL (II) ION, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-02-11 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 2: Synthesis and structure-activity relationships of benzothiazine-substituted quinolinediones
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2XP1
 
 | | Structure of the tandem SH2 domains from Antonospora locustae transcription elongation factor Spt6 | | Descriptor: | CHLORIDE ION, SPT6, SULFATE ION | | Authors: | Diebold, M.-L, Koch, M, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Noncanonical Tandem Sh2 Enables Interaction of Elongation Factor Spt6 with RNA Polymerase II.
J.Biol.Chem., 285, 2010
|
|
5VZ8
 
 | | Post-catalytic complex of human Polymerase Mu (G433A) mutant with incoming UTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5VZE
 
 | | Post-catalytic complex of human Polymerase Mu (W434A) mutant with incoming UTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
2I1R
 
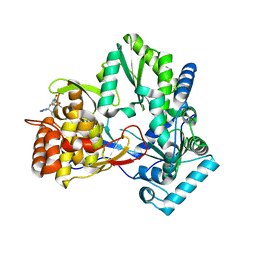 | | Novel Thiazolones as HCV NS5B Polymerase Inhibitors: Further Designs, Synthesis, SAR and X-ray Complex Structure | | Descriptor: | (5Z)-5-[(5-ETHYL-2-FURYL)METHYLENE]-2-{[(S)-(4-FLUOROPHENYL)(1H-TETRAZOL-5-YL)METHYL]AMINO}-1,3-THIAZOL-4(5H)-ONE, RNA-directed RNA polymerase (NS5B) (P68) | | Authors: | Yao, N, Yan, S. | | Deposit date: | 2006-08-14 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel thiazolones as HCV NS5B polymerase allosteric inhibitors: Further designs, SAR, and X-ray complex structure.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3Q0Z
 
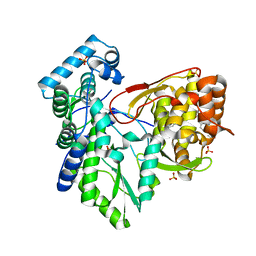 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5h-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2010-12-16 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Syntheses and initial evaluation of a series of indolo-fused heterocyclic inhibitors of the polymerase enzyme (NS5B) of the hepatitis C virus.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5N2E
 
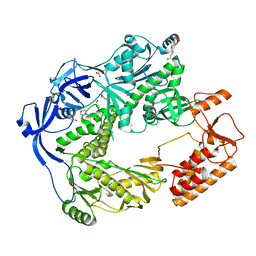 | | Structure of the E9 DNA polymerase from vaccinia virus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
1VYI
 
 | | Structure of the c-terminal domain of the polymerase cofactor of rabies virus: insights in function and evolution. | | Descriptor: | GLYCEROL, RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Mavrakis, M, McCarthy, A.A, Roche, S, Blondel, D, Ruigrok, R.W.H. | | Deposit date: | 2004-04-30 | | Release date: | 2004-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Function of the C-Terminal Domain of the Polymerase Cofactor of Rabies Virus
J.Mol.Biol., 343, 2004
|
|
5N2G
 
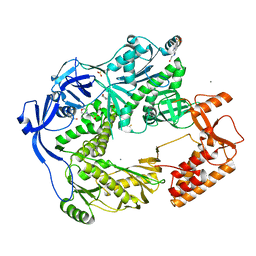 | | Structure of the E9 DNA polymerase from vaccinia virus in complex with manganese | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
2XWY
 
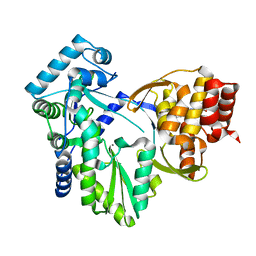 | | Structure of MK-3281, a Potent Non-Nucleoside Finger-Loop Inhibitor, in complex with the Hepatitis C Virus NS5B Polymerase | | Descriptor: | (7R)-14-cyclohexyl-7-{[2-(dimethylamino)ethyl](methyl)amino}-7,8-dihydro-6H-indolo[1,2-e][1,5]benzoxazocine-11-carboxylic acid, MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Di Marco, S, Baiocco, P. | | Deposit date: | 2010-11-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of (7R)-14-Cyclohexyl-7-{[2-(Dimethylamino)Ethyl] (Methyl)Amino}-7,8-Dihydro-6H-Indolo[1,2-E][1,5] Benzoxazocine -11-Carboxylic Acid (Mk-3281), a Potent and Orally Bioavailable Finger-Loop Inhibitor of the Hepatitis C Virus Ns5B Polymerase
J.Med.Chem., 54, 2011
|
|
5N2H
 
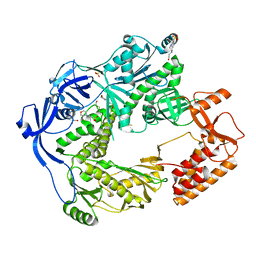 | | Structure of the E9 DNA polymerase exonuclease deficient mutant (D166A+E168A) from vaccinia virus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
2B43
 
 | |
1MNX
 
 | |
2EUL
 
 | | Structure of the transcription factor Gfh1. | | Descriptor: | ZINC ION, anti-cleavage anti-GreA transcription factor Gfh1 | | Authors: | Symersky, J, Perederina, A, Vassylyeva, M.N, Svetlov, V, Artsimovitch, I, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-28 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation through the RNA Polymerase Secondary Channel: STRUCTURAL AND FUNCTIONAL VARIABILITY OF THE COILED-COIL TRANSCRIPTION FACTORS.
J.Biol.Chem., 281, 2006
|
|
3I5K
 
 | | Crystal structure of the NS5B polymerase from Hepatitis C Virus (HCV) strain JFH1 | | Descriptor: | PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Simister, P.C, Schmitt, M, Lohmann, V, Bressanelli, S. | | Deposit date: | 2009-07-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of hepatitis C virus strain JFH1 polymerase
J.Virol., 83, 2009
|
|
7M0N
 
 | | The crystal structure of wild type PA endonuclease (A/Vietnam/1203/2004) in complex with Raltegravir | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Yun, M.K, Dubois, R, Rankovic, Z, White, S.W. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
