1NTX
 
 | |
1SMD
 
 | | HUMAN SALIVARY AMYLASE | | Descriptor: | AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Ramasubbu, N. | | Deposit date: | 1996-01-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human salivary alpha-amylase at 1.6 A resolution: implications for its role in the oral cavity.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2BSZ
 
 | | Structure of Mesorhizobium loti arylamine N-acetyltransferase 1 | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE 1 | | Authors: | Holton, S.J, Dairou, J, Sandy, J, Rodrigues-Lima, F, Dupret, J.-M, Noble, M.E.M, Sim, E. | | Deposit date: | 2005-05-24 | | Release date: | 2005-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Mesorhizobium Loti Arylamine N-Acetyltransferase 1.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
7N87
 
 | |
5ZWS
 
 | |
1CNX
 
 | |
1CNY
 
 | |
1CNW
 
 | |
2HDH
 
 | |
6S7O
 
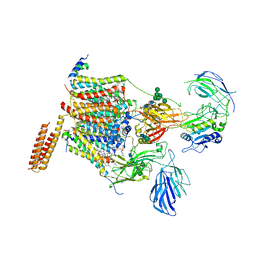 | | Cryo-EM structure of human oligosaccharyltransferase complex OST-A | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R},3~{R},4~{R},5'~{S},6~{S},7~{R},8~{S},9~{R},12~{R},13~{R},15~{S},16~{S},18~{R})-5',7,9,13-tetramethyl-3,15-bis(oxidanyl)spiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-16-yl]oxy-oxane-3,4,5-triol, (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit, ... | | Authors: | Ramirez, A.S, Kowal, J, Locher, K.P. | | Deposit date: | 2019-07-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B.
Science, 366, 2019
|
|
2CLX
 
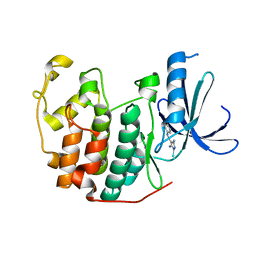 | | 4-Arylazo-3,5-diamino-1H-pyrazole CDK Inhibitors: SAR Study, Crystal Structure in Complex with CDK2, Selectivity, and Cellular Effects | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Krystof, V, Cankar, P, Frysova, I, Slouka, J, Kontopidis, G, Dzubak, P, Hajduch, M, Deazevedo, W.F, Paprskarova, M, Orsag, M, Rolcik, J, Latr, A, Fischer, P.M, Strnad, M. | | Deposit date: | 2006-05-02 | | Release date: | 2006-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 4-Arylazo-3,5-Diamino-1H-Pyrazole Cdk Inhibitors: Sar Study, Crystal Structure in Complex with Cdk2, Selectivity, and Cellular Effects
J.Med.Chem., 49, 2006
|
|
4YJ6
 
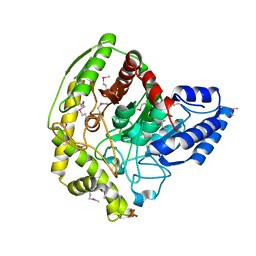 | | The Crystal Structure of a Bacterial Aryl Acylamidase Belonging to the Amidase signature (AS) enzymes family | | Descriptor: | Aryl acylamidase, PHOSPHATE ION | | Authors: | Lee, S, Park, E.-H, Ko, H.-J, Bang, W.-G, Choi, I.-G. | | Deposit date: | 2015-03-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Biochem.Biophys.Res.Commun., 467, 2015
|
|
3WJ9
 
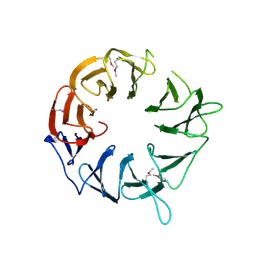 | |
4I6W
 
 | | 3-hydroxy-3-methylglutaryl (HMG) Coenzyme-A reductase complexed with thiomevalonate | | Descriptor: | (3S)-3-hydroxy-3-methyl-5-sulfanylpentanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, ... | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Burgner II, J.W, Rodwell, V.W, Wrensford, L.V, Critchelow, C.J, Min, J. | | Deposit date: | 2012-11-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
6RM3
 
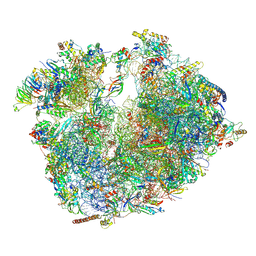 | | Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Barandun, J, Hunziker, M, Vossbrinck, C.R, Klinge, S. | | Deposit date: | 2019-05-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome.
Nat Microbiol, 4, 2019
|
|
3MPJ
 
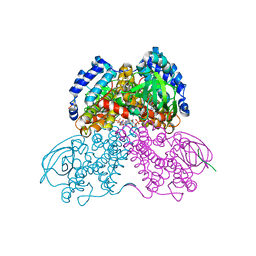 | | Structure of the glutaryl-coenzyme A dehydrogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, ... | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
3L1S
 
 | |
4P07
 
 | |
3IXL
 
 | |
4M4X
 
 | |
4FD4
 
 | |
4FD5
 
 | |
4FD7
 
 | | Crystal structure of insect putative arylalkylamine N-Acetyltransferase 7 from the yellow fever mosquito Aedes aegypt | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, SULFATE ION, ... | | Authors: | Han, Q, Robinson, R, Li, J. | | Deposit date: | 2012-05-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of insect arylalkylamine N-acetyltransferases: structural evidence from the yellow fever mosquito, Aedes aegypti.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4MC3
 
 | | Hedycaryol synthase in complex with Nerolidol | | Descriptor: | (3R,6E)-3,7,11-trimethyldodeca-1,6,10-trien-3-ol, Putative sesquiterpene cyclase | | Authors: | Baer, P, Rabe, P, Cirton, C, Oliveira Mann, C, Kaufmann, N, Groll, M, Dickschat, J. | | Deposit date: | 2013-08-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hedycaryol synthase in complex with nerolidol reveals terpene cyclase mechanism.
Chembiochem, 15, 2014
|
|
2VLB
 
 | | Structure of unliganded arylmalonate decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, ARYLMALONATE DECARBOXYLASE, BETA-MERCAPTOETHANOL, ... | | Authors: | Kuettner, E.B, Keim, A, Kircher, M, Rosmus, S, Strater, N. | | Deposit date: | 2008-01-11 | | Release date: | 2008-03-18 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Active Site Mobility Revealed by the Crystal Structure of Arylmalonate Decarboxylase from Bordetella Bronchiseptica
J.Mol.Biol., 377, 2008
|
|
