1H8X
 
 | | Domain-swapped Dimer of a Human Pancreatic Ribonuclease Variant | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Canals, A, Pous, J, Guasch, A, Benito, A, Ribo, M, Vilanova, M, Coll, M. | | Deposit date: | 2001-02-16 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of an Engineered Domain-Swapped Ribonuclease Dimer and its Implications for the Evolution of Proteins Toward Oligomerization
Structure, 9, 2001
|
|
1H1H
 
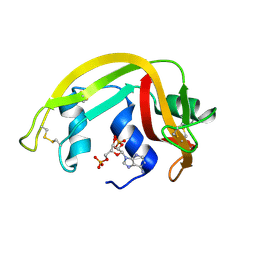 | | Crystal Structure of Eosinophil Cationic Protein in Complex with 2',5'-ADP at 2.0 A resolution Reveals the Details of the Ribonucleolytic Active site | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL CATIONIC PROTEIN | | Authors: | Mohan, C.G, Boix, E, Evans, H.R, Nikolovski, Z, Nogues, M.V, Cuchillo, C.M, Acharya, K.R. | | Deposit date: | 2002-07-15 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Eosinophil Cationic Protein in Complex with 2'5'-Adp at 2.0 A Resolution Reveals the Details of the Ribonucleolytic Active Site
Biochemistry, 41, 2002
|
|
1GV7
 
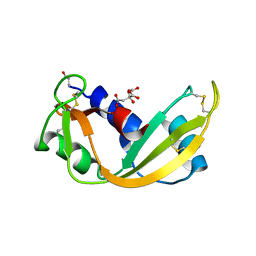 | | ARH-I, an angiogenin/RNase A chimera | | Descriptor: | ANGIOGENIN, CITRIC ACID | | Authors: | Holloway, D.E, Shapiro, R, Hares, M.C, Leonidas, D.D, Acharya, K.R. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Guest-Host Crosstalk in an Angiogenin-RNase A Chimeric Protein
Biochemistry, 41, 2002
|
|
1HBY
 
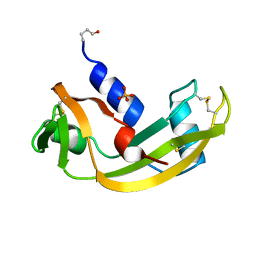 | | Binding of Phosphate and Pyrophosphate ions at the active site of human angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, PHOSPHATE ION | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.S, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-04-21 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
1HI3
 
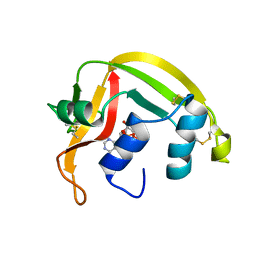 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine 2'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI4
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosien-3'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI2
 
 | | Eosinophil-derived Neurotoxin (EDN) - Sulphate Complex | | Descriptor: | EOSINOPHIL-DERIVED NEUROTOXIN, SULFATE ION | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI5
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
4OXF
 
 | |
4OKF
 
 | | RNase S in complex with an artificial peptide | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Genz, M, Strater, N. | | Deposit date: | 2014-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | X-ray structure of a RNase S variant in complex with an artificial peptide
To be Published
|
|
4OOH
 
 | |
4OWZ
 
 | |
4OXB
 
 | |
4OT4
 
 | |
4POU
 
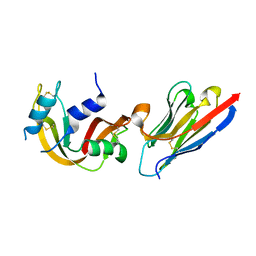 | | VHH-metal in Complex with RNase A | | Descriptor: | Ribonuclease pancreatic, VHH Antibody | | Authors: | Fanning, S.W, Walter, R, Horn, J.R. | | Deposit date: | 2014-02-26 | | Release date: | 2014-09-24 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of an engineered dual-specific antibody: conformational diversity leads to a hypervariable loop metal-binding site.
Protein Eng.Des.Sel., 27, 2014
|
|
4QFI
 
 | | The crystal structure of rat angiogenin-heparin complex | | Descriptor: | ACETIC ACID, Angiogenin, ZINC ION | | Authors: | Yeo, K.J, Hwang, E, Min, K.M, Hwang, K.Y, Jeon, Y.H, Chang, S.I, Cheong, H.K. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | The crystal structure of rat angiogenin-heparin complex
To be Published
|
|
4O36
 
 | | Semisynthetic RNase S1-15-H7/11-Q10 | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, S-peptide, ... | | Authors: | Genz, M, Strater, N. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | An Artificial Imine Reductase based on the Ribonuclease S scaffold
Chem.Cat.Chem, 2014
|
|
4O37
 
 | | seminsynthetic RNase S1-15-3Pl-7/11 | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, S-peptide, ... | | Authors: | Genz, M, Strater, N. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Artificial Imine Reductase based on the Ribonuclease S Scaffold
Chem.Cat.Chem, 2014
|
|
4QH3
 
 | |
4QFJ
 
 | | The crystal structure of rat angiogenin-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Angiogenin, ... | | Authors: | Yeo, K.J, Hwang, E, Min, K.M, Hwang, K.Y, Jeon, Y.H, Chang, S.I, Cheong, H.K. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | The crystal structure of rat angiogenin-heparin complex
To be Published
|
|
4RAT
 
 | |
4SRN
 
 | | STRUCTURAL CHANGES THAT ACCOMPANY THE REDUCED CATALYTIC EFFICIENCY OF TWO SEMISYNTHETIC RIBONUCLEASE ANALOGS | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | deMel, V.S.J, Martin, P.D, Doscher, M.S, Edwards, B.F.P. | | Deposit date: | 1991-05-20 | | Release date: | 1994-12-20 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural changes that accompany the reduced catalytic efficiency of two semisynthetic ribonuclease analogs.
J.Biol.Chem., 267, 1992
|
|
7PNP
 
 | | Human Angiogenin mutant-S28A | | Descriptor: | Angiogenin | | Authors: | Papaioannou, O.S.E, Leonidas, D.D. | | Deposit date: | 2021-09-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of the Human Angiogenin-Proliferating Cell Nuclear Antigen Interaction.
Biochemistry, 2023
|
|
2BZZ
 
 | | Crystal Structures of Eosinophil-derived Neurotoxin in Complex with the Inhibitors 5'-ATP, Ap3A, Ap4A and Ap5A | | Descriptor: | ACETIC ACID, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, NONSECRETORY RIBONUCLEASE | | Authors: | Baker, M.D, Holloway, D.E, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystal Structures of Eosinophil-Derived Neurotoxin (Edn) in Complex with the Inhibitors 5'- ATP, Ap(3)A, Ap(4)A, and Ap(5)A.
Biochemistry, 45, 2006
|
|
4J68
 
 | |
