3C22
 
 | |
3P7F
 
 | | Structure of the human Langerin carbohydrate recognition domain | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION | | Authors: | Skerra, A, Schiefner, A. | | Deposit date: | 2010-10-12 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The carbohydrate recognition domain of Langerin reveals high structural similarity with the one of DC-SIGN but an additional, calcium-independent sugar-binding site.
Mol.Immunol., 45, 2008
|
|
3M9Z
 
 | | Crystal Structure of extracellular domain of mouse NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | Authors: | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | Deposit date: | 2010-03-23 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular architecture of mouse activating NKR-P1 receptors.
J.Struct.Biol., 175, 2011
|
|
3CCK
 
 | | Human CD69 | | Descriptor: | CHLORIDE ION, Early activation antigen CD69 | | Authors: | Brynda, J, Vanek, O, Rezacova, P. | | Deposit date: | 2008-02-26 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Soluble recombinant CD69 receptors optimized to have an exceptional physical and chemical stability display prolonged circulation and remain intact in the blood of mice
Febs J., 275, 2008
|
|
6LKR
 
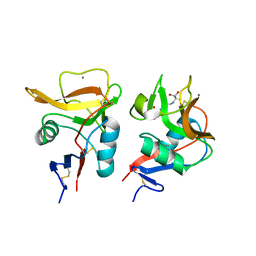 | | Crystal structure of mouse DCAR2 CRD domain complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, C-type lectin domain family 4, member b1, ... | | Authors: | Omahdi, Z, Horikawa, Y, Toyonaga, K, Kakuta, Y, Yamasaki, S. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the recognition of pathogen-derived phosphoglycolipids by C-type lectin receptor DCAR.
J.Biol.Chem., 295, 2020
|
|
1BUU
 
 | |
3PBF
 
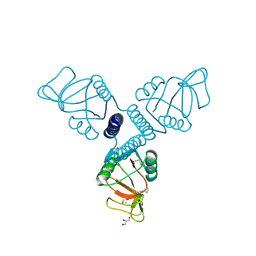 | | Surfactant Protein-A neck and carbohydrate recognition domain (NCRD) complexed with glycerol | | Descriptor: | CALCIUM ION, GLYCEROL, Pulmonary surfactant-associated protein A | | Authors: | Shang, F, Rynkiewicz, M.J, McCormack, F.X, Wu, H, Cafarella, T.M, Head, J, Seaton, B.A. | | Deposit date: | 2010-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic complexes of surfactant protein A and carbohydrates reveal ligand-induced conformational change.
J.Biol.Chem., 286, 2011
|
|
3ALU
 
 | | Crystal structure of CEL-IV complexed with Raffinose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Lectin CEL-IV, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3ALT
 
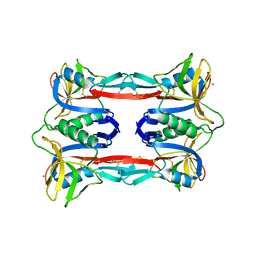 | | Crystal structure of CEL-IV complexed with Melibiose | | Descriptor: | CALCIUM ION, Lectin CEL-IV, C-type, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3ALS
 
 | | Crystal structure of CEL-IV | | Descriptor: | CALCIUM ION, Lectin CEL-IV, C-type | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3CAD
 
 | | Crystal structure of Natural Killer Cell Receptor, Ly49G | | Descriptor: | Lectin-related NK cell receptor LY49G1 | | Authors: | Cho, S. | | Deposit date: | 2008-02-19 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture of the Major Histocompatibility Complex Class I-binding Site of Ly49 Natural Killer Cell Receptors.
J.Biol.Chem., 283, 2008
|
|
1AFB
 
 | |
3VLG
 
 | | Crystal structure of the W150A mutant LOX-1 CTLD showing impaired OxLDL binding | | Descriptor: | Oxidized low-density lipoprotein receptor 1 | | Authors: | Nakano, S, Sugihara, M, Yamada, R, Katayanagi, K, Tate, S. | | Deposit date: | 2011-12-01 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural implication for the impaired binding of W150A mutant LOX-1 to oxidized low density lipoprotein, OxLDL
Biochim.Biophys.Acta, 1824, 2012
|
|
1DV8
 
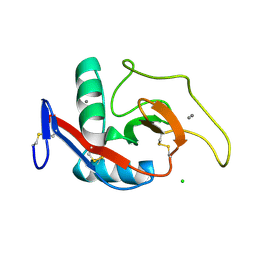 | | CRYSTAL STRUCTURE OF THE CARBOHYDRATE RECOGNITION DOMAIN OF THE H1 SUBUNIT OF THE ASIALOGLYCOPROTEIN RECEPTOR | | Descriptor: | ASIALOGLYCOPROTEIN RECEPTOR 1, CALCIUM ION, CHLORIDE ION | | Authors: | Meier, M, Bider, M.D, Malashkevich, V.N, Spiess, M, Burkhard, P. | | Deposit date: | 2000-01-20 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the carbohydrate recognition domain of the H1 subunit of the asialoglycoprotein receptor.
J.Mol.Biol., 300, 2000
|
|
4AK8
 
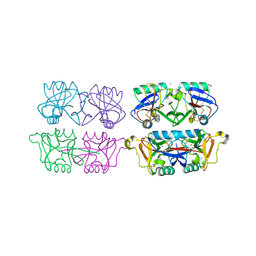 | | Structure of F241L mutant of langerin carbohydrate recognition domain. | | Descriptor: | C-TYPE LECTIN DOMAIN FAMILY 4 MEMBER K, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chabrol, E, Thepaut, M, Dezutter-Dambuyant, C, Vives, C, Marcoux, J, Kahn, R, Valadeau-Guilemond, J, Vachette, P, Durand, D, Fieschi, F. | | Deposit date: | 2012-02-22 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Alteration of the Langerin Oligomerization State Affects Birbeck Granule Formation.
Biophys.J., 108, 2015
|
|
3WH2
 
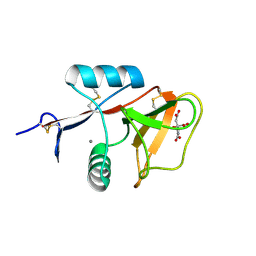 | | Human Mincle in complex with citrate | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION, CITRATE ANION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WHD
 
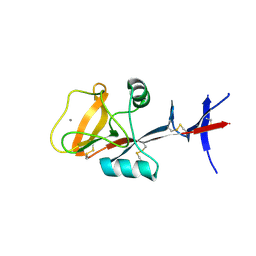 | | C-type lectin, human MCL | | Descriptor: | C-type lectin domain family 4 member D, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2ZIB
 
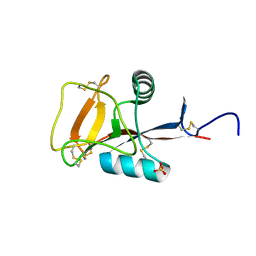 | | Crystal structure analysis of calcium-independent type II antifreeze protein | | Descriptor: | SULFATE ION, Type II antifreeze protein | | Authors: | Nishimiya, Y, Sato, R, Kondo, H, Noro, N, Sugimoto, H, Suzuki, M, Tsuda, S. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure and mutational analysis of Ca2+-independent type II antifreeze protein from longsnout poacher, Brachyopsis rostratus
J.Mol.Biol., 382, 2008
|
|
3WWK
 
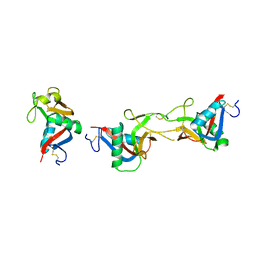 | | Crystal structure of CLEC-2 in complex with rhodocytin | | Descriptor: | C-type lectin domain family 1 member B, Snaclec rhodocytin subunit alpha, Snaclec rhodocytin subunit beta | | Authors: | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | Deposit date: | 2014-06-20 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
3WBP
 
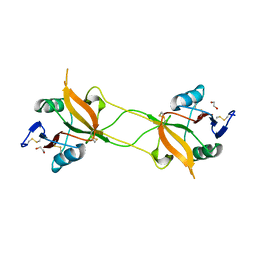 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-1) | | Descriptor: | 1,2-ETHANEDIOL, C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WBR
 
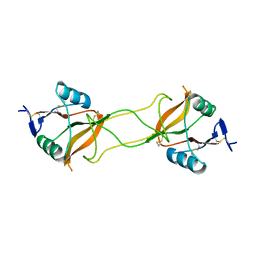 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-3) | | Descriptor: | C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WH3
 
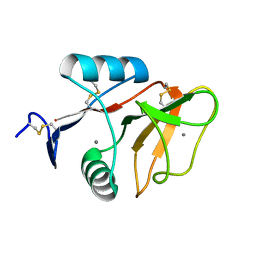 | | human Mincle, ligand free form | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WSR
 
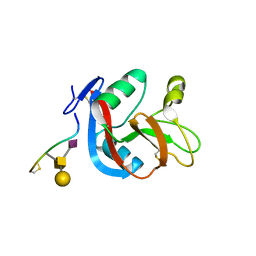 | | Crystal structure of CLEC-2 in complex with O-glycosylated podoplanin | | Descriptor: | C-type lectin domain family 1 member B, Peptide from Podoplanin, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | Deposit date: | 2014-03-20 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
1EGI
 
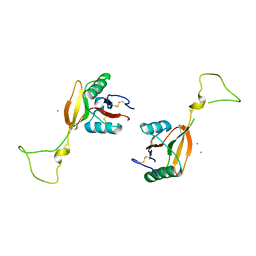 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
1EGG
 
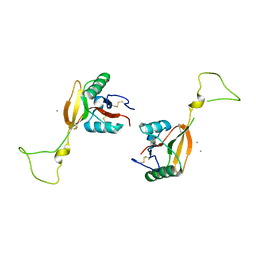 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
