4TW3
 
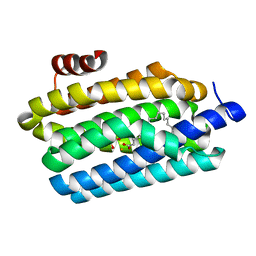 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
7NC2
 
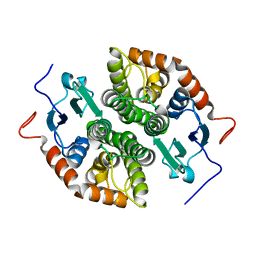 | |
7NCE
 
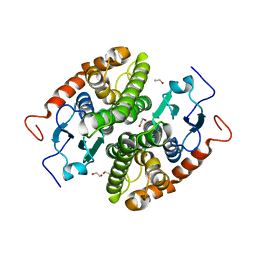 | | Glutathione-S-transferase GliG mutant N27A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4TWH
 
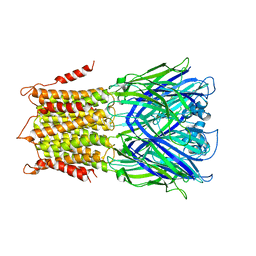 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant F16'S | | Descriptor: | Cys-loop ligand-gated ion channel | | Authors: | Ulens, C, Spurny, R, Thompson, A.J, Alqazzaz, M, Debaveye, S, Lu, H, Price, K, Villalgordo, J.M, Tresadern, G, Lynch, J.W, Lummis, S.C.R. | | Deposit date: | 2014-06-30 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Prokaryote Ligand-Gated Ion Channel ELIC Captured in a Pore Blocker-Bound Conformation by the Alzheimer's Disease Drug Memantine.
Structure, 22, 2014
|
|
7NCL
 
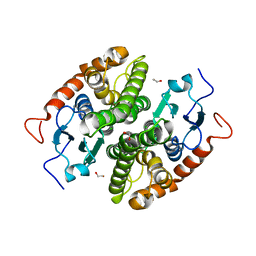 | | Glutathione-S-transferase GliG mutant E82Q | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC1
 
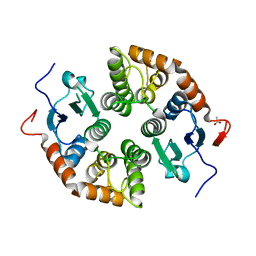 | |
4TYA
 
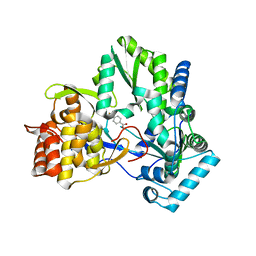 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 4-(trifluoromethyl)benzoic acid, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
7NCD
 
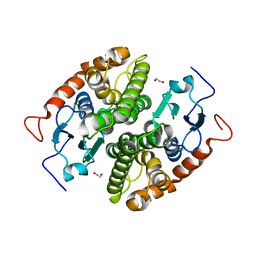 | | Glutathione-S-transferase GliG mutant N27D | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC8
 
 | | Glutathione-S-transferase GliG mutant S24A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4TWD
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with memantine | | Descriptor: | Cys-loop ligand-gated ion channel, Memantine | | Authors: | Ulens, C, Spurny, R, Thompson, A.J, Alqazzaz, M, Debaveye, S, Lu, H, Price, K, Villalgordo, J.M, Tresadern, G, Lynch, J.W, Lummis, S.C.R. | | Deposit date: | 2014-06-30 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Prokaryote Ligand-Gated Ion Channel ELIC Captured in a Pore Blocker-Bound Conformation by the Alzheimer's Disease Drug Memantine.
Structure, 22, 2014
|
|
7NC6
 
 | | Glutathione-S-transferase GliG in complex with cyclo[L-Phe-L-Ser]-bis-glutathione-adduct | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-3-[(2~{R},5~{R})-5-(hydroxymethyl)-3,6-bis(oxidanylidene)-2-(phenylmethyl)-5-sulfanyl-piperazin-2-yl]sulfanyl-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCO
 
 | | Glutathione-S-transferase GliG mutant K127R | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC5
 
 | |
4TXQ
 
 | | Crystal structure of LIP5 N-terminal domain complexed with CHMP1B MIM | | Descriptor: | Charged multivesicular body protein 1b, Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Vild, C.J, Xu, Z. | | Deposit date: | 2014-07-04 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Novel Mechanism of Regulating the ATPase VPS4 by Its Cofactor LIP5 and the Endosomal Sorting Complex Required for Transport (ESCRT)-III Protein CHMP5.
J.Biol.Chem., 290, 2015
|
|
7NCN
 
 | | Glutathione-S-transferase GliG mutant S83A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4U4P
 
 | | Crystal structure of the human condensin SMC hinge domain heterodimer with short coiled coils | | Descriptor: | Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Uchiyama, S, Kawahara, K, Hosokawa, Y, Fukakusa, S, Oki, H, Nakamura, S, Noda, M, Takino, R, Miyahara, Y, Maruno, T, Kobayashi, Y, Ohkubo, T, Fukui, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for dimer information and DNA recognition of human SMC proteins
to be published
|
|
7NC3
 
 | |
4TXW
 
 | | Crystal structure of CBM32-4 from the Clostridium perfringens NagH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Hyaluronoglucosaminidase | | Authors: | Grondin, J.M, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-07-07 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution Structure and Dynamics of Full-length GH84A, a multimodular B-N-acetylglucosaminidase from Clostridium perfringens
To Be Published
|
|
7NCP
 
 | | Glutathione-S-transferase GliG mutant K127A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG, SODIUM ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4TY1
 
 | |
7NDH
 
 | | Crystal structure of ZC3H12C PIN domain | | Descriptor: | 1,2-ETHANEDIOL, Probable ribonuclease ZC3H12C, SODIUM ION | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
4TYB
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (2R)-morpholin-4-yl(phenyl)ethanenitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
7NDI
 
 | | Crystal structure of ZC3H12C PIN domain with Mg2+ Ion | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable ribonuclease ZC3H12C, ... | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
7NPX
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 3-(3-Methoxyquinoxalin-2-yl)propanoic acid at 24 hours of soaking | | Descriptor: | 3-(3-methoxyquinoxalin-2-yl)propanoic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2021-02-28 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
7NDK
 
 | | Crystal structure of ZC3H12C PIN catalytic mutant | | Descriptor: | Probable ribonuclease ZC3H12C, SODIUM ION | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
