1CEL
 
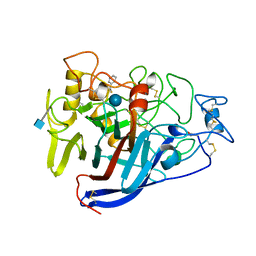 | |
5DLW
 
 | | Crystal structure of Autotaxin (ENPP2) with tauroursodeoxycholic acid (TUDCA) and lysophosphatidic acid (LPA) | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 2-{[(3alpha,5beta,7alpha,8alpha,14beta,17alpha)-3,7-dihydroxy-24-oxocholan-24-yl]amino}ethanesulfonic acid, CALCIUM ION, ... | | Authors: | Keune, W.J, Heidebrecht, T, Joosten, R.P, Perrakis, A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Steroid binding to Autotaxin links bile salts and lysophosphatidic acid signalling.
Nat Commun, 7, 2016
|
|
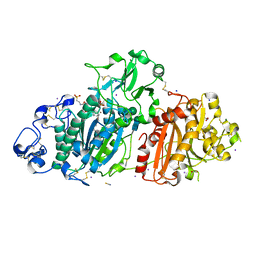
6HVJ
 
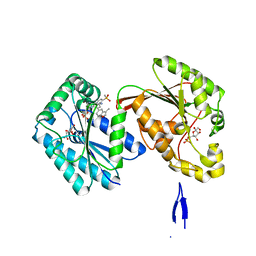 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 3 | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, 8-(3-methyl-1-benzofuran-5-yl)-~{N}-(4-methylsulfonylpyridin-3-yl)quinoxalin-6-amine, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery and Structure-Activity Relationships of N-Aryl 6-Aminoquinoxalines as Potent PFKFB3 Kinase Inhibitors.
ChemMedChem, 14, 2019
|
|
3WAX
 
 | | Crystal Structure of Autotaxin in Complex with 3BoA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
3WAY
 
 | | Crystal Structure of Autotaxin in Complex with 4BoA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
3WAV
 
 | | Crystal Structure of Autotaxin in Complex with Compound 10 | | Descriptor: | (5Z)-5-(3,4-dichlorobenzylidene)-2-(4-methylpiperazin-1-yl)-1,3-thiazol-4(5H)-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
6CVB
 
 | |
4UAF
 
 | |
4TVE
 
 | |
4TWI
 
 | | The structure of Sir2Af1 bound to a succinylated histone peptide | | Descriptor: | GLYCEROL, NAD-dependent protein deacylase 1, Succinylated H4 Peptide (aa8-20), ... | | Authors: | Ringel, A.E, Roman, C, Wolberger, C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Protein Sci., 23, 2014
|
|
4TXD
 
 | | Crystal structure of Thermofilum pendens Csc2 | | Descriptor: | Csc2, ZINC ION | | Authors: | Hrle, A, Conti, E. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structural analyses of the CRISPR protein Csc2 reveal the RNA-binding interface of the type I-D Cas7 family.
Rna Biol., 11, 2014
|
|
4TY9
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 5-(trifluoromethyl)pyridin-2-amine, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TZF
 
 | | Structure of metallo-beta lactamase | | Descriptor: | NDM-8 metallo-beta-lactamase, ZINC ION | | Authors: | Ferguson, J.A, Makena, A, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-07-10 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | structure of metallo-beta lactamase
To Be Published
|
|
4TZS
 
 | |
4TUM
 
 | | Crystal structure of Ankyrin Repeat Domain of AKR2 | | Descriptor: | Ankyrin repeat domain-containing protein 2 | | Authors: | Gwon, G.H, Cho, Y. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Ankyrin Repeat Domain of AKR2 Drives Chloroplast Targeting through Coincident Binding of Two Chloroplast Lipids.
Dev.Cell, 30, 2014
|
|
4U02
 
 | | Crystal structure of apo-TTHA1159 | | Descriptor: | Amino acid ABC transporter, ATP-binding protein, SULFATE ION | | Authors: | Karthiga Devi, S, Chichili, V.P.R, Velmurugan, D, Sivaraman, J. | | Deposit date: | 2014-07-11 | | Release date: | 2015-05-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for the hydrolysis of ATP by a nucleotide binding subunit of an amino acid ABC transporter from Thermus thermophilus
J.Struct.Biol., 190, 2015
|
|
6OIL
 
 | | Crystal structure of human VISTA extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, V-type immunoglobulin domain-containing suppressor of T-cell activation | | Authors: | Mehta, N, Cochran, J.R, Mathews, I.I. | | Deposit date: | 2019-04-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Functional Binding Epitope of V-domain Ig Suppressor of T Cell Activation.
Cell Rep, 28, 2019
|
|
4TUY
 
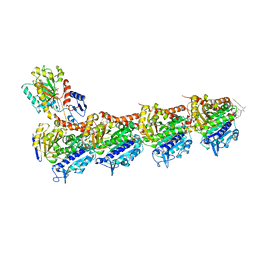 | | Tubulin-Rhizoxin complex | | Descriptor: | (1R,2R,3E,5R,7R,8S,10S,13E,16R)-8-hydroxy-10-[(2S,3R,4E,6E,8E)-3-methoxy-4,8-dimethyl-9-(2-methyl-1,3-oxazol-4-yl)nona-4,6,8-trien-2-yl]-2,7-dimethyl-6,11,19-trioxatricyclo[14.3.1.0~5,7~]icosa-3,13-diene-12,18-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Bargsten, K, Diaz, J.F, Marsh, M, Cuevas, C, Liniger, M, Neuhaus, C, Andreu, J.M, Altmann, K.H, Steinmetz, M.O. | | Deposit date: | 2014-06-25 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7NCB
 
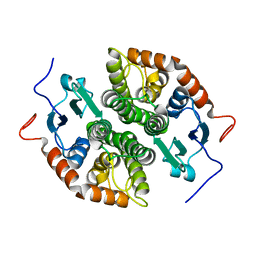 | | Glutathione-S-transferase GliG mutant H26A | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4TVT
 
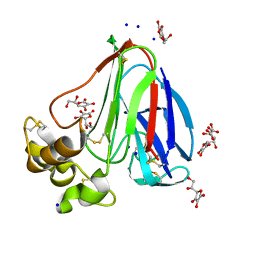 | | New ligand for thaumatin discovered using acoustic high throughput screening | | Descriptor: | ASCORBIC ACID, L(+)-TARTARIC ACID, SODIUM ION, ... | | Authors: | Teplitsky, E, Joshi, K, Ericson, D.L, Scalia, A, Mullen, J.D, Sweet, R.M, Soares, A.S. | | Deposit date: | 2014-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density.
J.Struct.Biol., 191, 2015
|
|
7NC9
 
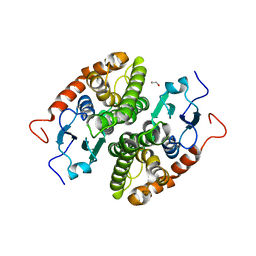 | | Glutathione-S-transferase GliG mutant H26N | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCM
 
 | | Glutathione-S-transferase GliG mutant E82A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4TVY
 
 | | Apo resorufin ligase | | Descriptor: | 5-(3,7-dihydroxy-10H-phenoxazin-2-yl)pentanamide, Lipoate-protein ligase A | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2014-06-28 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Computational design of a red fluorophore ligase for site-specific protein labeling in living cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7NCU
 
 | |
7NCT
 
 | | Glutathione-S-transferase GliG mutant K127G | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
