1EGD
 
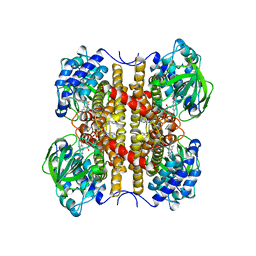 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1I7B
 
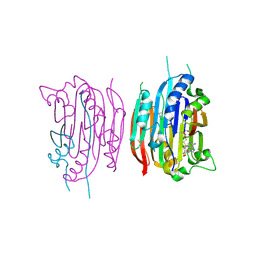 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND S-ADENOSYLMETHIONINE METHYL ESTER | | Descriptor: | 1,4-DIAMINOBUTANE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, S-ADENOSYLMETHIONINE DECARBOXYLASE BETA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1EME
 
 | |
1I7N
 
 | |
1ELX
 
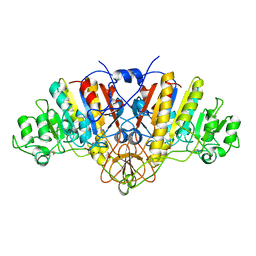 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102A) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
1EGC
 
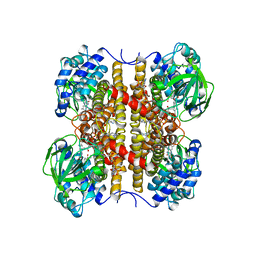 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE COMPLEXED WITH OCTANOYL-COA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE, OCTANOYL-COENZYME A | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1IAN
 
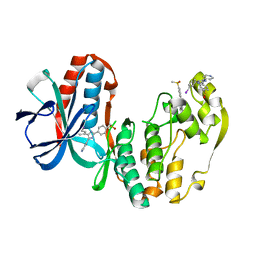 | | HUMAN P38 MAP KINASE INHIBITOR COMPLEX | | Descriptor: | 4-[5-(3-IODO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-1H-IMIDAZOL-4-YL]-PYRIDINE, P38 MAP KINASE | | Authors: | Tong, L. | | Deposit date: | 1997-03-07 | | Release date: | 1998-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A highly specific inhibitor of human p38 MAP kinase binds in the ATP pocket.
Nat.Struct.Biol., 4, 1997
|
|
1EFN
 
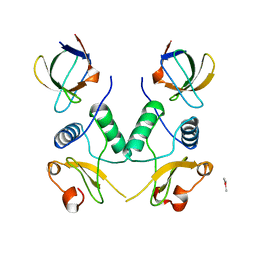 | |
1IUS
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE AT PH 5.0 | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1IVO
 
 | | Crystal Structure of the Complex of Human Epidermal Growth Factor and Receptor Extracellular Domains. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal Growth Factor Receptor, ... | | Authors: | Ogiso, H, Ishitani, R, Nureki, O, Fukai, S, Yamanaka, M, Kim, J.H, Saito, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-28 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Complex of Human Epidermal Growth Factor and Receptor Extracellular Domains.
Cell(Cambridge,Mass.), 110, 2002
|
|
1IFA
 
 | | THREE-DIMENSIONAL CRYSTAL STRUCTURE OF RECOMBINANT MURINE INTERFERON-BETA | | Descriptor: | ASPARAGINE, INTERFERON-BETA | | Authors: | Mitsui, Y, Senda, T, Matsuda, S, Kawano, G, Nakamura, K.T, Shimizu, H. | | Deposit date: | 1991-10-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional crystal structure of recombinant murine interferon-beta.
EMBO J., 11, 1992
|
|
1E6Y
 
 | | Methyl-coenzyme M reductase from Methanosarcina barkeri | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | Authors: | Grabarse, W, Ermler, U. | | Deposit date: | 2000-08-23 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparison of Three Methyl-Coenzyme M Reductases from Phylogenetically Distant Organisms: Unusual Amino Acid Modification, Conservation and Adaptation
J.Mol.Biol., 303, 2000
|
|
1IFH
 
 | |
1EAH
 
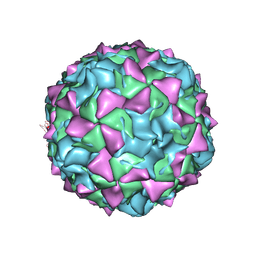 | | PV2L COMPLEXED WITH ANTIVIRAL AGENT SCH48973 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, POLIOVIRUS TYPE 2 COAT PROTEINS VP1 TO VP4 | | Authors: | Lentz, K, Arnold, E. | | Deposit date: | 1997-07-22 | | Release date: | 1998-09-16 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes.
Structure, 5, 1997
|
|
1ECG
 
 | | DON INACTIVATED ESCHERICHIA COLI GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE (PRPP) AMIDOTRANSFERASE | | Descriptor: | 5-OXO-L-NORLEUCINE, GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE AMIDOTRANSFERASE, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Krahn, J.M. | | Deposit date: | 1996-04-23 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site.
J.Biol.Chem., 271, 1996
|
|
1EUH
 
 | | APO FORM OF A NADP DEPENDENT ALDEHYDE DEHYDROGENASE FROM STREPTOCOCCUS MUTANS | | Descriptor: | NADP DEPENDENT NON PHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1998-11-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 290, 1999
|
|
1ECC
 
 | |
1IZ9
 
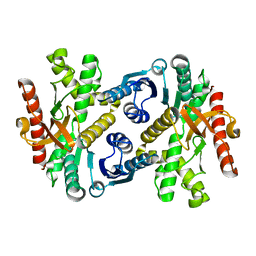 | | Crystal Structure of Malate Dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | MALATE DEHYDROGENASE | | Authors: | Hirose, R, Hasegawa, T, Yamano, A, Kuramitsu, S, Hamada, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Malate Dehydrogenase from Thermus themrophilus HB8
To be published
|
|
1ECB
 
 | |
1IUC
 
 | | Fucose-specific lectin from Aleuria aurantia with three ligands | | Descriptor: | Fucose-specific lectin, SULFATE ION, alpha-L-fucopyranose, ... | | Authors: | Fujihashi, M, Peapus, D.H, Kamiya, N, Nagata, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-01 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of Fucose-Specific Lectin from Aleuria aurantia Binding Ligands at Three of Its Five Sugar Recognition Sites
Biochemistry, 42, 2003
|
|
1E8C
 
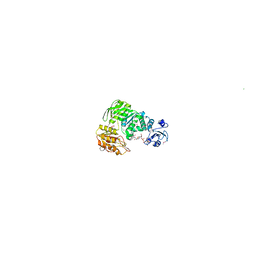 | | Structure of MurE the UDP-N-acetylmuramyl tripeptide synthetase from E. coli | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CHLORIDE ION, UDP-N-ACETYLMURAMOYLALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, ... | | Authors: | Gordon, E.J, Chantala, L, Dideberg, O. | | Deposit date: | 2000-09-19 | | Release date: | 2001-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Udp-N-Acetylmuramoyl-L-Alanyl-D-Glutamate: Meso-Diaminopimelate Ligase from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
1EUI
 
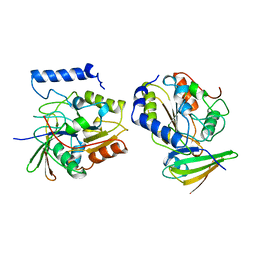 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Authors: | Ravishankar, R, Sagar, M.B, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray analysis of a complex of Escherichia coli uracil DNA glycosylase (EcUDG) with a proteinaceous inhibitor. The structure elucidation of a prokaryotic UDG.
Nucleic Acids Res., 26, 1998
|
|
1IUP
 
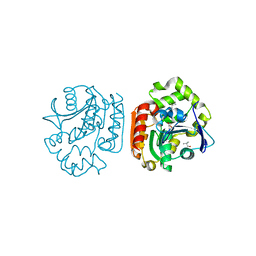 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant complexed with isobutyrates | | Descriptor: | 2-METHYL-PROPIONIC ACID, meta-Cleavage product hydrolase | | Authors: | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
1E5M
 
 | |
1E8W
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
