8BZL
 
 | | Human 20S Proteasome in complex with peptide activator peptide BLM42 | | Descriptor: | ARG-SER-TYR-TYR-SER, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Henneberg, F, Chari, A, Jankowska, E, Witkowska, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Peptidic, Blm10-based activators of human 20S proteasome in vitro and in cellulo enhance degradation of proteins connected with neurodegeneration.
To Be Published
|
|
8CNZ
 
 | | mmLarE-[4Fe-4S] phased by Fe-SAD | | Descriptor: | CHLORIDE ION, IODIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Pecqueur, L, Zecchin, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-02-24 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
8C6R
 
 | | PBP AccA from A. tumefaciens Bo542 in apoform 4 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CB9
 
 | | PBP AccA from A. tumefaciens Bo542 in complex with D-Glucose-2-phosphate | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-01-25 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
4QHX
 
 | |
4QGP
 
 | |
4QHB
 
 | |
4QNI
 
 | |
4QPV
 
 | |
4QT9
 
 | |
4R4G
 
 | |
4R03
 
 | |
4RCT
 
 | | Crystal structure of R-protein of NgoAVII restriction endonuclease | | Descriptor: | Restriction endonuclease R.NgoVII | | Authors: | Tamulaitiene, G, Silanskas, A, Grazulis, S, Zaremba, M, Siksnys, V. | | Deposit date: | 2014-09-17 | | Release date: | 2014-12-24 | | Last modified: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII.
Nucleic Acids Res., 42, 2014
|
|
4R4K
 
 | | Crystal structure of a cystatin-like protein (BACCAC_01506) from Bacteroides caccae ATCC 43185 at 1.69 A resolution | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of a hypothetical protein (BACCAC_01506) from Bacteroides caccae ATCC 43185 at 1.69 A resolution
To be published
|
|
4R7S
 
 | |
8D8I
 
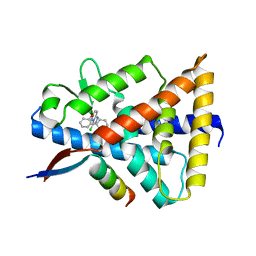 | | Crystal structure of Reverb alpha in complex with synthetic agonist | | Descriptor: | (4S)-6-[([1,1'-biphenyl]-2-yl)oxy]-3-chloro[1,2,4]triazolo[4,3-b]pyridazine, Nuclear receptor corepressor 1, Nuclear receptor subfamily 1 group D member 1 | | Authors: | Ronin, C, Ciesielski, F, Hegazy, L, Burris, P.T. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis of synthetic agonist activation of the nuclear receptor REV-ERB.
Nat Commun, 13, 2022
|
|
4QY7
 
 | |
4R79
 
 | |
4RAA
 
 | |
4RDB
 
 | |
4R1K
 
 | |
4R5O
 
 | |
4RGL
 
 | |
4RHO
 
 | |
4POI
 
 | |
