2Z9A
 
 | |
2W27
 
 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS YKUI PROTEIN, WITH AN EAL DOMAIN, IN COMPLEX WITH SUBSTRATE C-DI-GMP AND CALCIUM | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, YKUI PROTEIN | | Authors: | Padavattan, S, AndERSON, W.F, Schirmer, T. | | Deposit date: | 2008-10-24 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Ykui and its Complex with Second Messenger C-Di-Gmp Suggests Catalytic Mechanism of Phosphodiester Bond Cleavage by Eal Domains.
J.Biol.Chem., 284, 2009
|
|
3DS3
 
 | |
3DS4
 
 | |
3O5N
 
 | | Tetrahydroquinoline carboxylates are potent inhibitors of the Shank PDZ domain, a putative target in autism disorders | | Descriptor: | (3aS,4R,9bR)-9-nitro-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-4,6-dicarboxylic acid, SH3 and multiple ankyrin repeat domains protein 3 | | Authors: | Saupe, J, Roske, Y, Schillinger, C, Kamdem, N, Radetzki, S, Diehl, A, Oschkinat, H, Krause, G, Heinemann, U, Rademann, J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery, structure-activity relationship studies, and crystal structure of nonpeptide inhibitors bound to the shank3 PDZ domain.
Chemmedchem, 6, 2011
|
|
6UAE
 
 | | Ketosteroid isomerase (C. testosteroni) with truncated & designed loop for precise positioning of a catalytic E38 | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, EQUILENIN, Ketosteroid isomerase with designed loop, ... | | Authors: | Krivacic, C, Kundert, K, Thompson, M.C, Fraser, J.S, Kortemme, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ketosteroid isomerase (C. testosteroni) with truncated & designed loop for precise positioning of a catalytic E38
To Be Published
|
|
5KCR
 
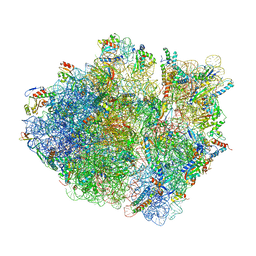 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Avilamycin C, mRNA and P-site tRNA at 3.6A resolution | | Descriptor: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5YMW
 
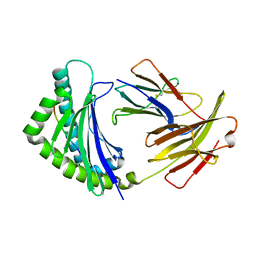 | | Crystal structure of 8-mer peptide from Rous sarcoma virus in complex with BF2*1201 | | Descriptor: | Beta-2-microglobulin, Class I histocompatibility antigen, F10 alpha chain, ... | | Authors: | Xiao, J, Xiang, W, Qi, J, Chai, Y, Liu, W.J, Gao, G.F. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | An Invariant Arginine in Common with MHC Class II Allows Extension at the C-Terminal End of Peptides Bound to Chicken MHC Class I.
J Immunol., 201, 2018
|
|
3DS1
 
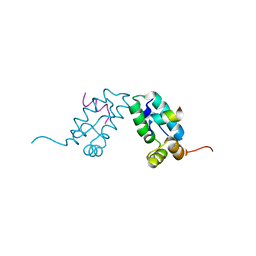 | |
3DS0
 
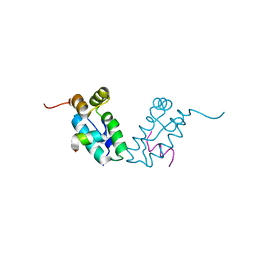 | |
3DS2
 
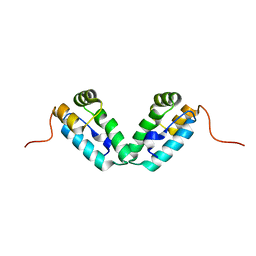 | | HIV-1 capsid C-terminal domain mutant (Y169A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Vaney, M.-C, Igonet, S, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
1E29
 
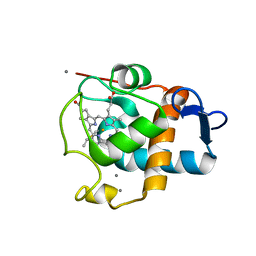 | | PSII associated cytochrome C549 from Synechocystis sp. | | Descriptor: | CALCIUM ION, CYTOCHROME C549, HEME C | | Authors: | Frazao, C, Enguita, F.J, Coelho, R, Sheldrick, G.M. | | Deposit date: | 2000-05-19 | | Release date: | 2001-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal Structure of Low-Potential Cytochrome C549 from Synechocystis Sp. Pcc 6803 at 1.21A Resolution
J.Biol.Inorg.Chem., 6, 2001
|
|
3DS5
 
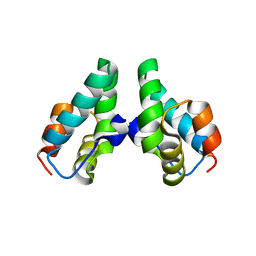 | | HIV-1 capsid C-terminal domain mutant (N183A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
3DTJ
 
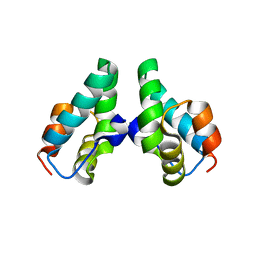 | | HIV-1 capsid C-terminal domain mutant (E187A) | | Descriptor: | HIV-1 capsid protein | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
3A4C
 
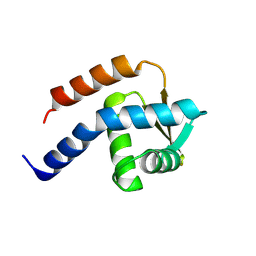 | | Crystal structure of cdt1 C terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Cho, Y, Lee, J.H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
1R5S
 
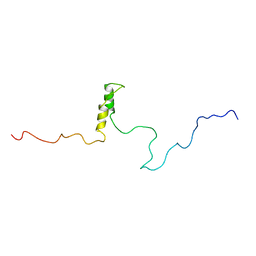 | | Connexin 43 Carboxyl Terminal Domain | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Sorgen, P.L, Duffy, H.S, Mario, D, Sahoo, P, Coombs, W, Delmar, M, Spray, D.C. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1
J.Biol.Chem., 279, 2004
|
|
5EPY
 
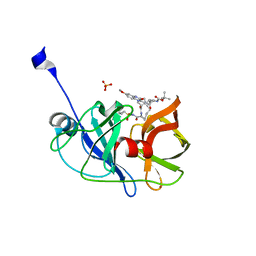 | | Crystal structure of HCV NS3/4A protease A156T variant in complex with 5172-mcP1P3 (MK-5172 P1-P3 macrocyclic analogue) | | Descriptor: | 2-Methyl-2-propanyl {(2R,6S,12Z,13aS,14aR,16aS)-14a-[(cyclopropylsulfonyl)carbamoyl]-2-[(3-ethyl-7-methoxy-2-quinoxalinyl)oxy]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclop ropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D.I, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
5EPN
 
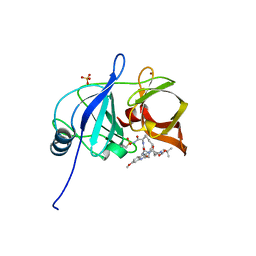 | | Crystal structure of HCV NS3/4A protease in complex with 5172-mcP1P3 (MK-5172 P1-P3 macrocyclic analogue) | | Descriptor: | 2-Methyl-2-propanyl {(2R,6S,12Z,13aS,14aR,16aS)-14a-[(cyclopropylsulfonyl)carbamoyl]-2-[(3-ethyl-7-methoxy-2-quinoxalinyl)oxy]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclop ropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D.I, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-11 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
5VP9
 
 | |
5HRV
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with 1-ethylisochromeno[3,4-c]pyrazol-5(2H)-one) compound | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylisochromeno[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1 | | Authors: | Tallant, C, Myrianthopoulos, V, Gaboriaud-Kolar, N, Newman, J.A, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Mikros, E, Knapp, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
6MYD
 
 | | Structure of zebrafish TRAF6 in complex with STING CTT | | Descriptor: | STING CTT, Transmembrane protein 173, SULFATE ION, ... | | Authors: | de Oliveira Mann, C.C, Orzalli, M.H, King, D.S, Kagan, J.C, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2018-11-01 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Modular Architecture of the STING C-Terminal Tail Allows Interferon and NF-kappa B Signaling Adaptation.
Cell Rep, 27, 2019
|
|
3T4R
 
 | |
1CYJ
 
 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
1YLG
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
3LW1
 
 | | Binary complex of 14-3-3 sigma and p53 pT387-peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Schumacher, B, Mondry, J, Thiel, P, Weyand, M, Ottmann, C. | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure of the p53 C-terminus bound to 14-3-3: Implications for stabilization of the p53 tetramer
Febs Lett., 584, 2010
|
|
