7PEH
 
 | | Crystal Structure of a Class D Carbapenemase | | Descriptor: | 1-BUTANOL, Beta-lactamase | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
8QLV
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 4 | | Descriptor: | Oligopeptide-binding protein AliB, VAL-MET-VAL-LYS-GLY-PRO-GLY-PRO-GLY-ARG | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
6EZL
 
 | |
5UDV
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with iron | | Descriptor: | FE (III) ION, Lactate racemization operon protein LarE, PHOSPHATE ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7KK5
 
 | |
8QZ4
 
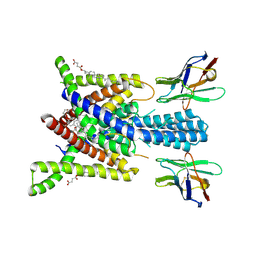 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb76) | | Descriptor: | BARIUM ION, CHOLESTEROL HEMISUCCINATE, Nanobody 76, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
6F08
 
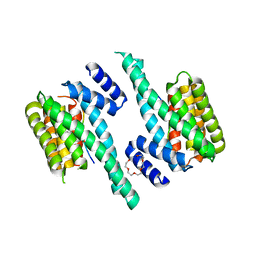 | | 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1) | | Descriptor: | 14-3-3 protein zeta/delta, PENTAETHYLENE GLYCOL, Son of sevenless homolog 1 | | Authors: | Ballone, A, Centorrino, F, Ottmann, C, Guo, S, Leysen, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1).
J. Struct. Biol., 202, 2018
|
|
6AXR
 
 | |
5U5L
 
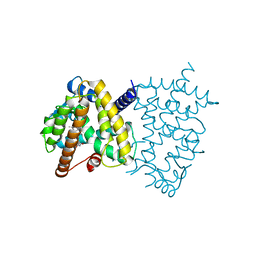 | | X-ray Crystal Structure of the PPARgamma Ligand Binding Domain in Complex with Rivoglitazone | | Descriptor: | (5S)-5-({4-[(6-methoxy-1-methyl-1H-benzimidazol-2-yl)methoxy]phenyl}methyl)-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Bruning, J.B, Rajapaksha, H, Wegener, K, Bhatia, H. | | Deposit date: | 2016-12-06 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | X-ray crystal structure of rivoglitazone bound to PPAR gamma and PPAR subtype selectivity of TZDs.
Biochim. Biophys. Acta, 1861, 2017
|
|
7KMI
 
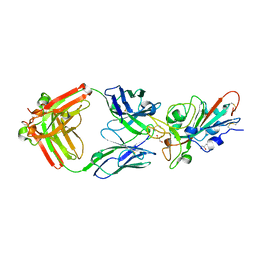 | | LY-CoV481 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LY-CoV481 Fab heavy chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
6J32
 
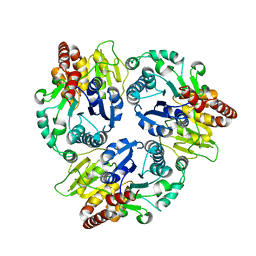 | | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin | | Descriptor: | Kcn28 | | Authors: | Shi, J, Liu, C.L, Zhang, B, Guo, W.J, Zhu, J.P, Xu, X, Xu, Q, Jiao, R.H, Tan, R.X, Ge, H.M. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Genome mining and biosynthesis of kitacinnamycins as a STING activator.
Chem Sci, 10, 2019
|
|
6F0Q
 
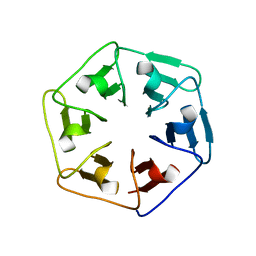 | |
6AXV
 
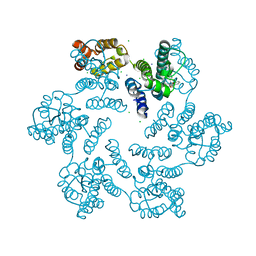 | |
6Y31
 
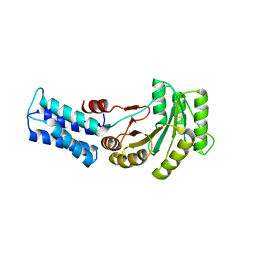 | | NG domain of human SRP54 T117 deletion mutant | | Descriptor: | Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Lapouge, K, Becker, M.M.M, Kotova, I, Haas, M, Carapito, R, Wild, K, Bahram, S, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
5U5W
 
 | | CcP gateless cavity | | Descriptor: | 3-methylquinolin-4-amine, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Fischer, M, Shoichet, B.K. | | Deposit date: | 2016-12-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8QOK
 
 | |
7KIV
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F. | | Deposit date: | 2020-10-24 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Structural Characterization of Diazabicyclooctane beta-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7ZT0
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | 1-(2-piperazin-1-ylethyl)-5-pyridin-4-yl-indole-2-carboxamide, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-05-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
8QOP
 
 | |
8A2V
 
 | |
8QOQ
 
 | |
6B1U
 
 | | Structure of full-length human AMPK (a2b1g1) in complex with a small molecule activator SC4 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
5F91
 
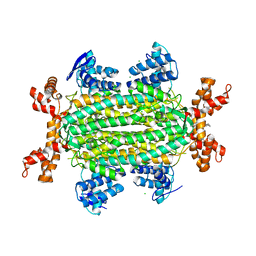 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator (N-(5-(azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fumarate hydratase class II, ... | | Authors: | Kasbekar, M, Fischer, G, Mott, B.T, Yasgar, A, Hyvonen, M, Boshoff, H.I, Abell, C, Barry, C.E, Thomas, C.J. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Selective small molecule inhibitor of the Mycobacterium tuberculosis fumarate hydratase reveals an allosteric regulatory site.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6Y3W
 
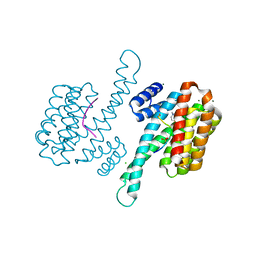 | | Ternary complex of 14-3-3 sigma (C38N), Estrogen Related Receptor gamma (DBD) phosphopeptide, and disulfide PPI stabilizer 1 | | Descriptor: | 14-3-3 protein sigma, 3-fluoranyl-~{N}-methyl-~{N}-(2-sulfanylethyl)benzamide, CHLORIDE ION, ... | | Authors: | Somsen, B.A, Ottmann, C. | | Deposit date: | 2020-02-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Fluorescence Anisotropy-Based Tethering for Discovery of Protein-Protein Interaction Stabilizers.
Acs Chem.Biol., 15, 2020
|
|
6AYK
 
 | |
