7FBY
 
 | | Crystal Structure of PH0140 from Pyrococcus horikosii OT3 | | Descriptor: | 1,2-ETHANEDIOL, ISOLEUCINE, Transcriptional regulatory protein | | Authors: | Richard, M, Ahmad, M, Pal, R.K, Biswal, B.K, Jeyakanthan, J. | | Deposit date: | 2021-07-13 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of PH0140 from Pyrococcus horikosii OT3
To Be Published
|
|
3DKE
 
 | | Polar and non-polar cavities in phage T4 lysozyme | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AZIDE ION, ... | | Authors: | Liu, L.J, Matthews, B.W. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Use of experimental crystallographic phases to examine the hydration of polar and nonpolar cavities in T4 lysozyme
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3DUX
 
 | | Understanding Thrombin Inhibition | | Descriptor: | 3-cyclohexyl-D-alanyl-N-(3-chlorobenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-07-18 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
3DV7
 
 | |
1RJK
 
 | | crystal structure of the rat vitamin D receptor ligand binding domain complexed with 2MD and a synthetic peptide containing the NR2 box of DRIP 205 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-2-METHYLENE-CYCLOHEXANE-1,3-DIO L, Peroxisome proliferator-activated receptor binding protein, Vitamin D3 receptor | | Authors: | Vanhooke, J.L, Benning, M.M, Bauer, C.B, Pike, J.W, DeLuca, H.F. | | Deposit date: | 2003-11-19 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular Structure of the Rat Vitamin D Receptor Ligand Binding Domain Complexed with 2-Carbon-Substituted Vitamin D(3) Hormone Analogues and a LXXLL-Containing Coactivator Peptide
Biochemistry, 43, 2004
|
|
1RNJ
 
 | | Crystal structure of inactive mutant dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
7FCR
 
 | |
8CQX
 
 | | Ribokinase from T.sp mutant A92G | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribokinase | | Authors: | Timofeev, V.I, Shevtsov, M.B, Abramchik, Y.A, Kostromina, M.A, Zayats, E.A, Kuranova, I.P, Esipov, R.S. | | Deposit date: | 2023-03-07 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Ribokinase from T.sp mutant A92G
To Be Published
|
|
7LBA
 
 | | E. coli Agmatinase | | Descriptor: | Agmatinase, MANGANESE (II) ION | | Authors: | Chitrakar, I, Ahmed, S.F, Torelli, A.T, French, J.B. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the E. coli agmatinase, SPEB.
Plos One, 16, 2021
|
|
7FCS
 
 | |
3DNA
 
 | |
7FCY
 
 | | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor | | Descriptor: | (1S)-2-methyl-1-(2-methyl-1,2,4-triazol-3-yl)propan-1-amine, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor
To Be Published
|
|
6RBL
 
 | | TETR(D) E147A MUTANT IN COMPLEX WITH DOXYCYCLINE AND MAGNESIUM | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hinrichs, W, Palm, G.J, Berndt, L, Girbardt, B. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
7FDO
 
 | |
5BS0
 
 | | MAGE-A3 Reactive TCR in complex with Titin Epitope in HLA-A1 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Raman, M.C.C, Rizkallah, P.J, Simmons, R, Donellan, Z, Dukes, J, Bossi, G, LeProvost, G, Mahon, T, Hickman, E, Lomax, M, Oates, J, Hassan, N, Vuidepot, A, Sami, M, Cole, D.K, Jakobsen, B.K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Direct molecular mimicry enables off-target cardiovascular toxicity by an enhanced affinity TCR designed for cancer immunotherapy.
Sci Rep, 6, 2016
|
|
3DRM
 
 | | 2.2 Angstrom Crystal Structure of Thr114Phe Alpha1-Antitrypsin | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Gooptu, B, Nobeli, I, Purkiss, A, Phillips, R.L, Mallya, M, Lomas, D.A, Barrett, T.E. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design.
J.Mol.Biol., 387, 2009
|
|
2OJX
 
 | | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization | | Descriptor: | Serine/threonine-protein kinase PLK1, Synthetic peptide | | Authors: | Garcia-Alvarez, B, de Carcer, G, Ibanez, S, Bragado-Nilsson, E, Montoya, G. | | Deposit date: | 2007-01-15 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7LHN
 
 | |
2OED
 
 | | GB3 solution structure obtained by refinement of X-ray structure with dipolar couplings | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ulmer, T.S, Ramirez, B.E, Delaglio, F, Bax, A, Grishaev, A. | | Deposit date: | 2006-12-29 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy
J.Am.Chem.Soc., 125, 2003
|
|
3DIK
 
 | |
5I9J
 
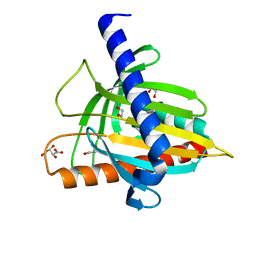 | | Structure of the cholesterol and lutein-binding domain of human STARD3 at 1.74A | | Descriptor: | 1,2-ETHANEDIOL, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Horvath, M.P, Bernstein, P.S, Li, B, George, E.W, Tran, Q.T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the lutein-binding domain of human StARD3 at 1.74 angstrom resolution and model of a complex with lutein.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7LHM
 
 | |
7LHO
 
 | |
2O4E
 
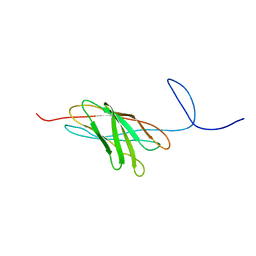 | | The solution structure of a protein-protein interaction module from a family 84 glycoside hydrolase of Clostridium perfringens | | Descriptor: | O-GlcNAcase nagJ | | Authors: | Chitayat, S, Adams, J.J, Gregg, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase.
J.Mol.Biol., 375, 2008
|
|
6R81
 
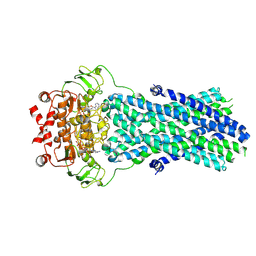 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP and Mg solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2019-03-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
