6EYT
 
 | |
6BZJ
 
 | | Solution structure of AGL55 | | Descriptor: | M protein | | Authors: | Qiu, C, Yuan, Y, Castellino, F.J. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
6F3N
 
 | |
6F4J
 
 | | Crystal structure of Drosophila melanogaster SNF/U2A' complex | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Probable U2 small nuclear ribonucleoprotein A', SULFATE ION, ... | | Authors: | Weber, G, DeKoster, G, Holton, N, Hall, K.B, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6BSC
 
 | |
6FAW
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2c-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-[2-(2-methoxyethoxy)phenyl]-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6F98
 
 | | Solution structure of the RING domain of the E3 ubiquitin ligase HRD1 | | Descriptor: | E3 ubiquitin-protein ligase, ZINC ION | | Authors: | Kniss, A, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RING domain of the E3 ubiquitin ligase HRD1
To Be Published
|
|
6F99
 
 | |
6BQI
 
 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 2020
|
|
6CDB
 
 | | Crystal Structure of V66L CzrA in the Zn(II)bound state | | Descriptor: | ArsR family transcriptional regulator, CHLORIDE ION, SODIUM ION, ... | | Authors: | Capdevila, D.A, Campanello, G, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional Role of Solvent Entropy and Conformational Entropy of Metal Binding in a Dynamically Driven Allosteric System.
J. Am. Chem. Soc., 140, 2018
|
|
6FBW
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2f-II with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{S})-(3-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6F2Q
 
 | | Neutron crystal structure of perdeuterated galectin-3C in the ligand-free form | | Descriptor: | Galectin-3 | | Authors: | Manzoni, F, Blakeley, M.P, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.03 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
6F3M
 
 | |
6COV
 
 | | CSP2-l14 | | Descriptor: | Competence-stimulating peptide type 2 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6F4I
 
 | | Crystal structure of Drosophila melanogaster SNF | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Weber, G, Holton, N, Hall, K.B, DeKoster, G, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6BYD
 
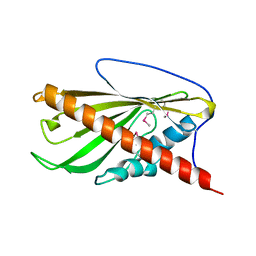 | | Crystal structure of the second StART domain of yeast Lam4 | | Descriptor: | Membrane-anchored lipid-binding protein LAM4 | | Authors: | Jentsch, J.A, Kiburu, I.N, Wu, J, Pandey, K, Boudker, O, Menon, A.K. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Structural basis of sterol binding and transport by a yeast StARkin domain.
J. Biol. Chem., 293, 2018
|
|
6C0M
 
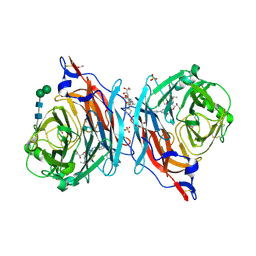 | | The synthesis, biological evaluation and structural insights of unsaturated 3-N-substituted sialic acids as probes of human parainfluenza virus-3 haemagglutinin-neuraminidase | | Descriptor: | 1,2-ETHANEDIOL, 2,6-anhydro-3,5-dideoxy-5-[(2-methylpropanoyl)amino]-3-(4-phenyl-1H-1,2,3-triazol-1-yl)-D-glycero-D-galacto-non-2-enoni c acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, Ve, T, von Itzstein, M. | | Deposit date: | 2018-01-01 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insights into Human Parainfluenza Virus 3 Hemagglutinin-Neuraminidase Using Unsaturated 3- N-Substituted Sialic Acids as Probes.
ACS Chem. Biol., 13, 2018
|
|
6C1J
 
 | | Crystal Structure of Ketosteroid Isomerase Y32F/Y57F/D40N mutant from Pseudomonas Putida (pKSI) bound to 3,4-dinitrophenol | | Descriptor: | 3,4-dinitrophenol, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yabukarski, F, Pinney, M.M, Herschlag, D. | | Deposit date: | 2018-01-04 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.063 Å) | | Cite: | Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein.
J. Am. Chem. Soc., 140, 2018
|
|
6FGN
 
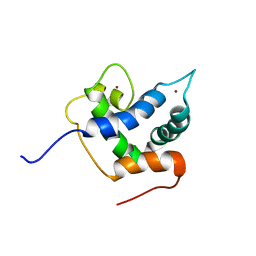 | | Solution Structure of p300Taz2-p63TA | | Descriptor: | Histone acetyltransferase p300,Tumor protein 63, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
6CVT
 
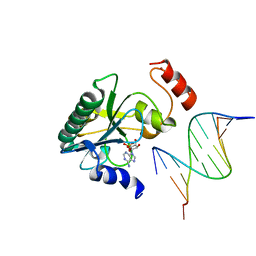 | | Human Aprataxin (Aptx) V263G bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6FPP
 
 | | Structure of S. pombe Mmi1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, YTH domain-containing protein mmi1 | | Authors: | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
6COO
 
 | | CSP1-E1A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6FQ2
 
 | | Structure of minimal sequence for left -handed G-quadruplex formation | | Descriptor: | DNA (5'-D(*GP*TP*GP*GP*TP*GP*GP*TP*GP*GP*TP*G)-3'), POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Heddi, B, Bakalar, B. | | Deposit date: | 2018-02-13 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6CRL
 
 | | HusA haemophore from Porphyromonas gingivalis | | Descriptor: | hypothetical protein PG_2227 | | Authors: | Gell, D.A, Kwan, A.H, Horne, J, Hugrass, B.M, Collins, D.A.T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural properties of a haemophore facilitate targeted elimination of the pathogen Porphyromonas gingivalis.
Nat Commun, 9, 2018
|
|
6FYH
 
 | | Disulfide between ubiquitin G76C and the E3 HECT ligase Huwe1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Jaeckl, M, Hartmann, M.D, Wiesner, S. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | beta-Sheet Augmentation Is a Conserved Mechanism of Priming HECT E3 Ligases for Ubiquitin Ligation.
J. Mol. Biol., 430, 2018
|
|
