7OKK
 
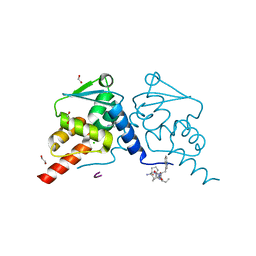 | | Crystal structure of human BCL6 BTB domain in complex with compound 12e | | Descriptor: | 1,2-ETHANEDIOL, 2-[[6-[(2-chloranyl-3-cyano-pyridin-4-yl)amino]-2-oxidanylidene-1H-quinolin-4-yl]amino]-N-methyl-ethanamide, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKE
 
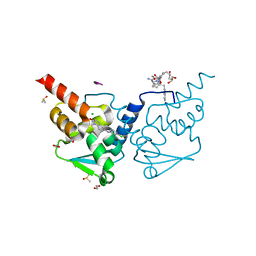 | | Crystal structure of human BCL6 BTB domain in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[(1-methyl-2-oxidanylidene-quinolin-6-yl)amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKI
 
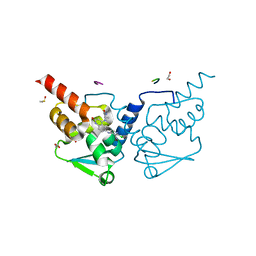 | | Crystal structure of human BCL6 BTB domain in complex with compound 12b | | Descriptor: | (2R)-2-[[6-[(2-chloranyl-3-cyano-pyridin-4-yl)amino]-2-oxidanylidene-1H-quinolin-4-yl]amino]-N-methyl-propanamide, 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKL
 
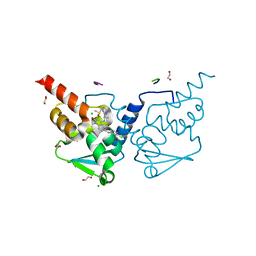 | | Crystal structure of human BCL6 BTB domain in complex with compound 13e | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[1-methyl-2-oxidanylidene-4-[[(1R)-1-pyrimidin-2-ylethyl]amino]quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
4IN9
 
 | | Structure of karilysin MMP-like catalytic domain in complex with inhibitory tetrapeptide SWFP | | Descriptor: | GLYCEROL, Karilysin protease, POTASSIUM ION, ... | | Authors: | Guevara, T, Ksiazek, M, Skottrup, P.D, Cerda-Costa, N, Trillo-Muyo, S, de Diego, I, Riise, E, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the catalytic domain of the Tannerella forsythia matrix metallopeptidase karilysin in complex with a tetrapeptidic inhibitor.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6A70
 
 | | Structure of the human PKD1/PKD2 complex | | Descriptor: | Polycystin-1, Polycystin-2 | | Authors: | Su, Q, Hu, F, Ge, X, Lei, J, Yu, S, Wang, T, Zhou, Q, Mei, C, Shi, Y. | | Deposit date: | 2018-06-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the human PKD1-PKD2 complex.
Science, 361, 2018
|
|
1YF6
 
 | | Structure of a quintuple mutant of photosynthetic reaction center from rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Paddock, M.L, Chang, C, Xu, Q, Abresch, E.C, Axelrod, H.L. | | Deposit date: | 2004-12-30 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Quinone (Q(B)) Reduction by B-Branch Electron Transfer in Mutant Bacterial Reaction Centers from Rhodobacter sphaeroides: Quantum Efficiency and X-ray Structure.
Biochemistry, 44, 2005
|
|
5XMW
 
 | | Selenomethionine-derivated ZHD | | Descriptor: | Zearalenone lactonase | | Authors: | Hu, X.J. | | Deposit date: | 2017-05-16 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
8DDF
 
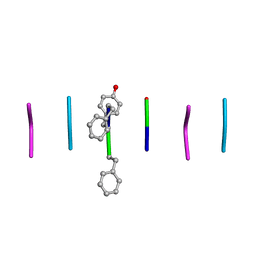 | | Quasi-racemic mixture of L-FWF and D-FYF peptide reveals rippled beta-sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, DPN-DTY-DPN, PHE-TRP-PHE | | Authors: | Sawaya, M.R, Hazari, A, Eisenberg, D.E. | | Deposit date: | 2022-06-18 | | Release date: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The rippled beta-sheet layer configuration-a novel supramolecular architecture based on predictions by Pauling and Corey.
Chem Sci, 13, 2022
|
|
6MJL
 
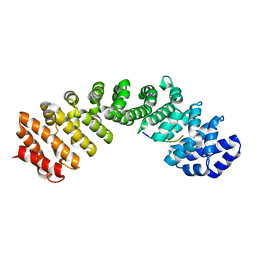 | | Crystal structure of ChREBP NLS peptide bound to importin alpha. | | Descriptor: | ChREBP Peptide ASN-TYR-TRP-LYS-ARG-ARG-ILE-GLU-VAL, Importin subunit alpha-1 | | Authors: | Jung, H, Uyeda, K. | | Deposit date: | 2018-09-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of importin alpha and the nuclear localization peptide of ChREBP, and small compound inhibitors of ChREBP-importin alpha interactions.
Biochem.J., 477, 2020
|
|
1E6D
 
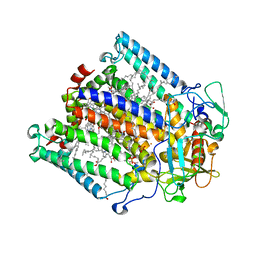 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH TRP M115 REPLACED WITH PHE (CHAIN M, WM115F) PHE M197 REPLACED WITH ARG (CHAIN M, FM197R) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Ridge, J.P, Fyfe, P.K, McAuley, K.E, Van Brederode, M.E, Robert, B, Van Grondelle, R, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 2000-08-11 | | Release date: | 2000-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Examination of How Structural Changes Can Affect the Rate of Electron Transfer in a Mutated Bacterial Photoreaction Centre
Biochem.J., 351, 2000
|
|
6A2B
 
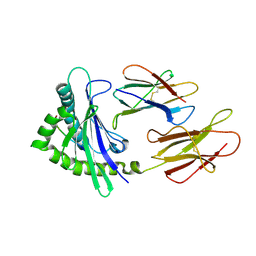 | | Crystal Structure of Xenopus laevis MHC I complex | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, TYR-MET-MET-PRO-ARG-HIS-TRP-PRO-ILE | | Authors: | Ma, L.Z, Xia, C. | | Deposit date: | 2018-06-10 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Glimpse of the Peptide Profile Presentation byXenopus laevisMHC Class I: Crystal Structure of pXela-UAA Reveals a Distinct Peptide-Binding Groove.
J Immunol., 204, 2020
|
|
6AMU
 
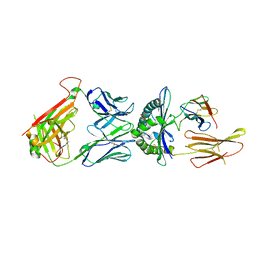 | |
6AMT
 
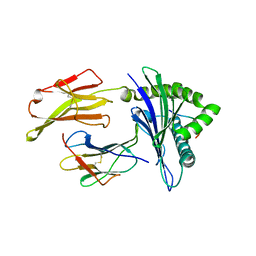 | |
1C8P
 
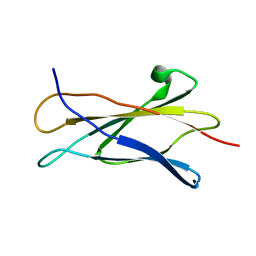 | | NMR STRUCTURE OF THE LIGAND BINDING DOMAIN OF THE COMMON BETA-CHAIN IN THE GM-CSF, IL-3 AND IL-5 RECEPTORS | | Descriptor: | CYTOKINE RECEPTOR COMMON BETA CHAIN | | Authors: | Mulhern, T.D, D'Andrea, R.J, Gaunt, C, Vandeleur, L, Vadas, M.A, Lopez, A.F, Booker, G.W, Bagley, C.J. | | Deposit date: | 1999-10-05 | | Release date: | 2000-06-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the cytokine-binding domain of the common beta-chain of the receptors for granulocyte-macrophage colony-stimulating factor, interleukin-3 and interleukin-5.
J.Mol.Biol., 297, 2000
|
|
4HTC
 
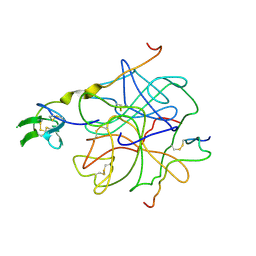 | | THE REFINED STRUCTURE OF THE HIRUDIN-THROMBIN COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | Tulinsky, A, Rydel, T.J, Bode, W, Huber, R. | | Deposit date: | 1993-06-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure of the hirudin-thrombin complex.
J.Mol.Biol., 221, 1991
|
|
7Q7S
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(ethylamino)-1,3-dimethyl-2-oxidanylidene-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7Q7T
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[(2S)-2,7-dimethyl-5,6-bis(oxidanylidene)-2,3-dihydro-1H-[1,4]oxazepino[6,5-c]quinolin-10-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
3HTC
 
 | | THE STRUCTURE OF A COMPLEX OF RECOMBINANT HIRUDIN AND HUMAN ALPHA-THROMBIN | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUDIN VARIANT 2 | | Authors: | Tulinsky, A, Rydel, T.J, Ravichandran, K.G, Huber, R, Bode, W. | | Deposit date: | 1993-06-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a complex of recombinant hirudin and human alpha-thrombin.
Science, 249, 1990
|
|
5C8Z
 
 | | ZHD-ZGR complex after ZHD crystal soaking in ZEN for 30min | | Descriptor: | 2,4-dihydroxy-6-[(1E,10S)-10-hydroxy-6-oxoundec-1-en-1-yl]benzoic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X.-J, Qi, Q, Yang, W.-J. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3DWJ
 
 | | Heme-proximal W188H mutant of inducible nitric oxide synthase | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, AMMONIUM ION, Nitric oxide synthase, ... | | Authors: | Tejero, J, Biswas, A, Wang, Z.-Q, Haque, M.M, Hemann, C, Zweier, J.L, Page, R.C, Misra, S, Stuehr, D.J. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Stabilization and Characterization of a Heme-Oxy Reaction Intermediate in Inducible Nitric-oxide Synthase
J.Biol.Chem., 283, 2008
|
|
7R7Y
 
 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | Authors: | Li, X.L, Ng, R, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7V
 
 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7W
 
 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, MHC class I antigen | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7X
 
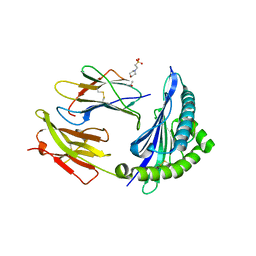 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
