3UVV
 
 | | Crystal Structure of the ligand binding domains of the thyroid receptor:retinoid X receptor complexed with 3,3',5 triiodo-L-thyronine and 9-cis retinoic acid | | Descriptor: | (9cis)-retinoic acid, 3,5,3'TRIIODOTHYRONINE, Retinoic acid receptor RXR-alpha, ... | | Authors: | Fernandez, E.J, Putcha, B.-D.K, Wright, E, Brunzelle, J.S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for negative cooperativity within agonist-bound TR:RXR heterodimers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1GOU
 
 | |
1EEF
 
 | | HEAT-LABILE ENTEROTOXIN B-PENTAMER COMPLEXED WITH BOUND LIGAND PEPG | | Descriptor: | 2-PHENETHYL-2,3-DIHYDRO-PHTHALAZINE-1,4-DIONE, PROTEIN (HEAT-LABILE ENTEROTOXIN B CHAIN), alpha-D-galactopyranose | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploration of the GM1 receptor-binding site of heat-labile enterotoxin and cholera toxin by phenyl-ring-containing galactose derivatives.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4K6Y
 
 | |
4E0U
 
 | | Crystal structure of CdpNPT in complex with thiolodiphosphate and (S)-benzodiazependione | | Descriptor: | (3S)-3-(1H-indol-3-ylmethyl)-3,4-dihydro-1H-1,4-benzodiazepine-2,5-dione, 1,2-ETHANEDIOL, Cyclic dipeptide N-prenyltransferase, ... | | Authors: | Schuller, J.M, Zocher, G, Stehle, T. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
6IJO
 
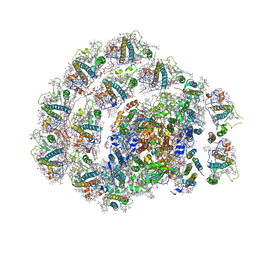 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
5KVP
 
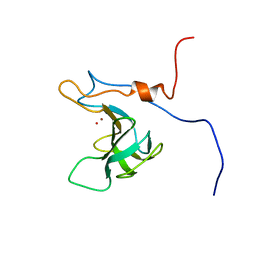 | |
4K72
 
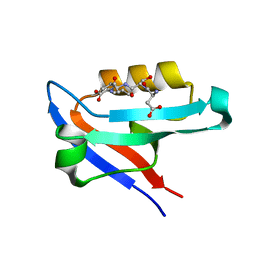 | |
1AZ4
 
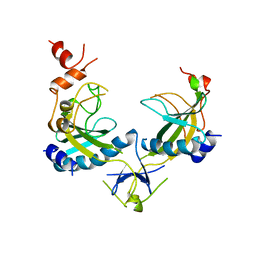 | | ECORV ENDONUCLEASE, UNLIGANDED, FORM B, T93A MUTANT | | Descriptor: | ECORV ENDONUCLEASE | | Authors: | Perona, J, Martin, A. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis.
J.Mol.Biol., 273, 1997
|
|
2OPM
 
 | | Human Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-461 | | Descriptor: | 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, Farnesyl pyrophosphate synthetase (FPP synthetase) (FPS) (Farnesyl diphosphate synthetase) [Includes: Dimethylallyltranstransferase (EC 2.5.1.1); Geranyltranstransferase (EC 2.5.1.10)], MAGNESIUM ION, ... | | Authors: | Cao, R, Gao, Y.G, Robinson, H, Goddard, A. | | Deposit date: | 2007-01-29 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
6J40
 
 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
3HL1
 
 | |
3HM4
 
 | |
6A1I
 
 | | Crystal structure of a synthase 1 from Santalum album | | Descriptor: | CARBONATE ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Han, X, Ko, T.P, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a synthase 1 from Santalum album in complex with ligand
To Be Published
|
|
2PCS
 
 | | Crystal structure of conserved protein from Geobacillus kaustophilus | | Descriptor: | Conserved protein, UNKNOWN LIGAND | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Wu, B, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of conserved protein from Geobacillus kaustophilus
To be Published
|
|
3KL7
 
 | |
6KHI
 
 | | Supercomplex for cylic electron transport in cyanobacteria | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
6KM9
 
 | | Crystal structure of SucA from Vibrio vulnificus | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Seo, P.W, Kim, J.S. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | Understanding the molecular properties of the E1 subunit (SucA) of alpha-ketoglutarate dehydrogenase complex from Vibrio vulnificus for the enantioselective ligation of acetaldehydes into (R)-acetoin.
Catalysis Science And Technology, 2020
|
|
6K3K
 
 | | Solution structure of APOBEC3G-CD2 with ssDNA, Product B | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, DNA/RNA (5'-D(*AP*TP*TP*CP*UP*(ICY)P*AP*AP*TP*T)-3'), ZINC ION | | Authors: | Cao, C, Yan, X, Lan, W, Wang, C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigations on the Interactions between Cytidine Deaminase Human APOBEC3G and DNA.
Chem Asian J, 14, 2019
|
|
6K3J
 
 | | Solution structure of APOBEC3G-CD2 with ssDNA, Product A | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*UP*(IUR)P*AP*AP*TP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Cao, C, Yan, X, Lan, W, Wang, C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigations on the Interactions between Cytidine Deaminase Human APOBEC3G and DNA.
Chem Asian J, 14, 2019
|
|
3L2N
 
 | |
6KWU
 
 | | A Crystal Structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Kiya, M, Makabe, K. | | Deposit date: | 2019-09-08 | | Release date: | 2020-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | beta-Strand-mediated Domain-swapping in the Absence of Hydrophobic Core Repacking.
J.Mol.Biol., 436, 2024
|
|
4NW4
 
 | |
2PY6
 
 | |
2Q83
 
 | |
