8GB1
 
 | | Crystal structure of SAMHD1 dimer bound to deoxyguanosine linked inhibitor | | Descriptor: | 5'-O-[(R)-(3-{[(1M)-3'-bromo[1,1'-biphenyl]-3-carbonyl]amino}propoxy)(hydroxy)phosphoryl]-2'-deoxyguanosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Deoxyguanosine-Linked Bifunctional Inhibitor of SAMHD1 dNTPase Activity and Nucleic Acid Binding.
Acs Chem.Biol., 18, 2023
|
|
3AAT
 
 | |
3ADK
 
 | |
1GWZ
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE PROTEIN TYROSINE PHOSPHATASE SHP-1 | | Descriptor: | SHP-1 | | Authors: | Yang, J, Liang, X, Niu, T, Meng, W, Zhao, Z, Zhou, G.W. | | Deposit date: | 1998-08-22 | | Release date: | 1999-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic domain of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 273, 1998
|
|
5ABZ
 
 | | Complex of the FimH lectin with a C-linked naphtyl alpha-D-mannoside in soaked trigonal crystals at 2.40 A resolution | | Descriptor: | (2R,3S,4R,5S,6R)-2-(HYDROXYMETHYL)-6-[3-(NAPHTHALEN-1-YL)PROPYL]OXANE-3,4,5-TRIOL, FIMH, NICKEL (II) ION | | Authors: | de Ruyck, J, Bouckaert, J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of C-Mannosylated Anti-Adhesives Bound to the Type 1 Fimbrial Fimh Adhesin
Iucrj, 3, 2016
|
|
1APZ
 
 | |
3UTB
 
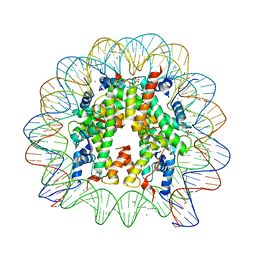 | | Crystal Structure of Nucleosome Core Particle Assembled with the 146b Alpha-Satellite Sequence (NCP146b) | | Descriptor: | 146-mer DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
3BUZ
 
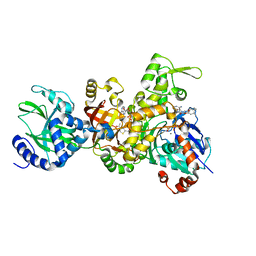 | | Crystal structure of ia-bTAD-actin complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Tsuge, H, Nagahama, M, Oda, M, Iwamoto, S, Utsunomiya, H, Marquez, V.E, Katunuma, N, Nishizawa, M, Sakurai, J. | | Deposit date: | 2008-01-04 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1W98
 
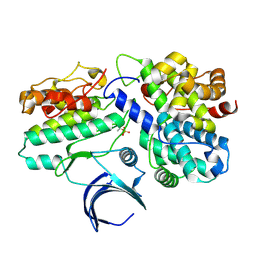 | | The structural basis of CDK2 activation by cyclin E | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, G1/S-SPECIFIC CYCLIN E1 | | Authors: | Lowe, E.D, Honda, R, Dubinina, E, Skamnaki, V, Cook, A, Johnson, L.N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of Cyclin E1/Cdk2: Implications for Cdk2 Activation and Cdk2-Independent Roles
Embo J., 24, 2005
|
|
5UWW
 
 | |
5UWJ
 
 | |
5AAL
 
 | | Complex of the FimH lectin with a C-linked para-biphenyl ethylene alpha-D-mannoside in soaked trigonal crystals at 2.45 A resolution | | Descriptor: | (2R,3S,4R,5S,6R)-2-(hydroxymethyl)-6-[(Z)-3-(4-phenylphenyl)prop-2-enyl]oxane-3,4,5-triol, FIMH, NICKEL (II) ION, ... | | Authors: | De Ruyck, J, Bouckaert, J. | | Deposit date: | 2015-07-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of C-Mannosylated Anti-Adhesives Bound to the Type 1 Fimbrial Fimh Adhesin
Iucrj, 3, 2016
|
|
3UTA
 
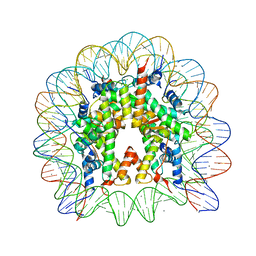 | | Crystal Structure of Nucleosome Core Particle Assembled with an Alpha-Satellite Sequence Containing Two TTAAA elements (NCP-TA2) | | Descriptor: | 145-mer DNA, CHLORIDE ION, Histone H2A, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
3UT9
 
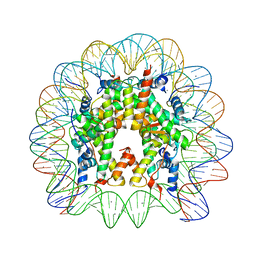 | | Crystal Structure of Nucleosome Core Particle Assembled with a Palindromic Widom '601' Derivative (NCP-601L) | | Descriptor: | 145-mer DNA, CHLORIDE ION, Histone H2A, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
5T94
 
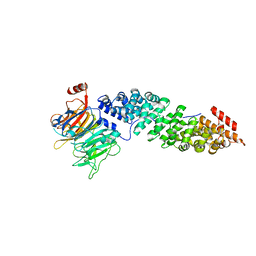 | | Crystal structure of Kap60 bound to yeast RCC1 (Prp20) | | Descriptor: | Guanine nucleotide exchange factor SRM1, Importin subunit alpha | | Authors: | Sankhala, R.S, Lokareddy, R.K, Pumroy, R.A, Cingolani, G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Three-dimensional context rather than NLS amino acid sequence determines importin alpha subtype specificity for RCC1.
Nat Commun, 8, 2017
|
|
3LUD
 
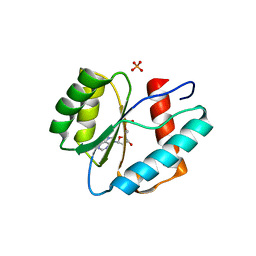 | |
3LUK
 
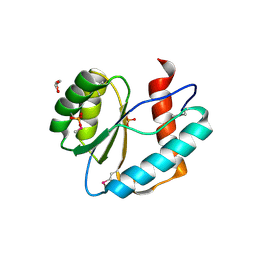 | |
5UQV
 
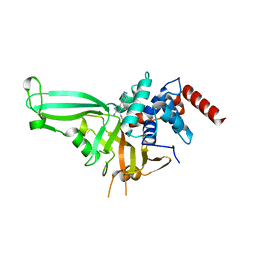 | |
5AAP
 
 | |
3BO0
 
 | | Ribosome-SecY complex | | Descriptor: | 23S RIBOSOMAL RNA, PREPROTEIN TRANSLOCASE SecE SUBUNIT, PREPROTEIN TRANSLOCASE SecY SUBUNIT, ... | | Authors: | Akey, C.W, Menetret, J.F. | | Deposit date: | 2007-12-15 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Ribosome binding of a single copy of the SecY complex: implications for protein translocation
Mol.Cell, 28, 2007
|
|
5UWR
 
 | |
6TIM
 
 | |
5UWU
 
 | |
3BQU
 
 | | Crystal Structure of the 2F5 Fab'-3H6 Fab Complex | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, 3H6 Fab heavy chain, ... | | Authors: | Bryson, S, Julien, J.-P, Isenman, D.E, Kunert, R, Katinger, H, Pai, E.F. | | Deposit date: | 2007-12-20 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the complex between the F(ab)' fragment of the cross-neutralizing anti-HIV-1 antibody 2F5 and the F(ab) fragment of its anti-idiotypic antibody 3H6.
J.Mol.Biol., 382, 2008
|
|
5UWI
 
 | |
