5E7W
 
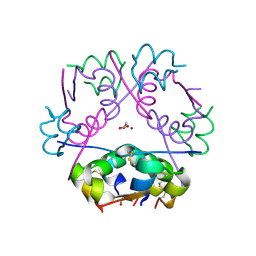 | | X-ray Structure of Human Recombinant 2Zn insulin at 0.92 Angstrom | | Descriptor: | ACETATE ION, Insulin, N-PROPANOL, ... | | Authors: | Lisgarten, D.R, Naylor, C.E, Palmer, R.A, Lobley, C.M.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.9519 Å) | | Cite: | Ultra-high resolution X-ray structures of two forms of human recombinant insulin at 100 K.
Chem Cent J, 11, 2017
|
|
6FW2
 
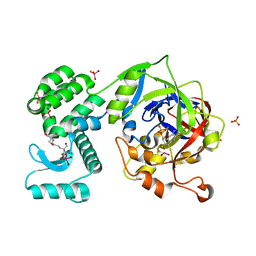 | | Crystal Structure of human mARC1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MOLYBDATE ION, Mitochondrial amidoxime-reducing component 1,Endolysin,Mitochondrial amidoxime-reducing component 1, ... | | Authors: | Kubitza, C, Scheidig, A. | | Deposit date: | 2018-03-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human mARC1 reveals its exceptional position among eukaryotic molybdenum enzymes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5U5O
 
 | |
7CYG
 
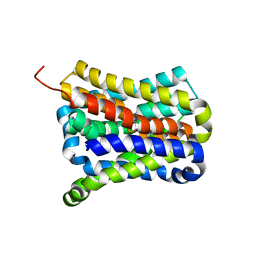 | | Crystal structure of a cysteine-pair mutant (Y113C-P190C) of a bacterial bile acid transporter before disulfide bond formation | | Descriptor: | Transporter, sodium/bile acid symporter family | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6Q0R
 
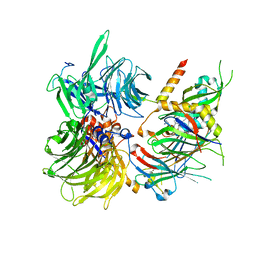 | | Structure of DDB1-DDA1-DCAF15 complex bound to E7820 and RBM39 | | Descriptor: | 3-cyano-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6FWD
 
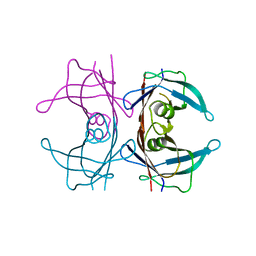 | |
6V8W
 
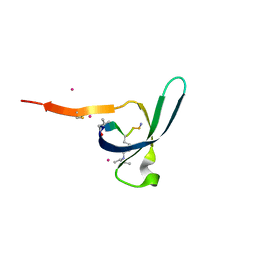 | | CDYL2 chromodomain in complex with a synthetic peptide | | Descriptor: | Chromodomain Y-like protein 2, IVA-PHE-ALA-PHE-5T3-SER-NH2, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, James, L.I, Lamb, K.N, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6XT6
 
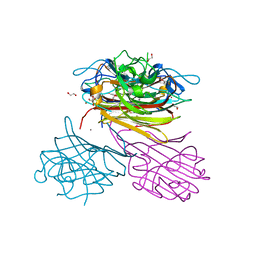 | | pro-concanavalin A: Precursor of circularly permuted concanavalin A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Concanavalin-A, ... | | Authors: | Nonis, S.G, Haywood, J, Schmidberger, J.W, Bond, C.S. | | Deposit date: | 2020-07-17 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural and biochemical analyses of concanavalin A circular permutation by jack bean asparaginyl endopeptidase.
Plant Cell, 33, 2021
|
|
6FWO
 
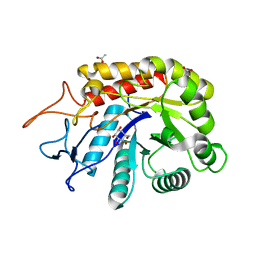 | | Structure of an E336Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-1,2-anhydro-mannose hydrolyzed by enzyme | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
6V98
 
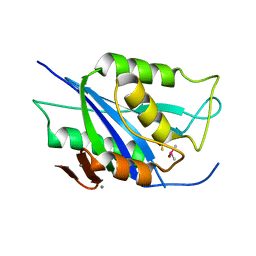 | | Crystal structure of Type VI secretion system effector, TseH (VCA0285) | | Descriptor: | CALCIUM ION, Cysteine hydrolase | | Authors: | Watanabe, N, Hersch, S.J, Dong, T.G, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Envelope stress responses defend against type six secretion system attacks independently of immunity proteins.
Nat Microbiol, 5, 2020
|
|
5VTE
 
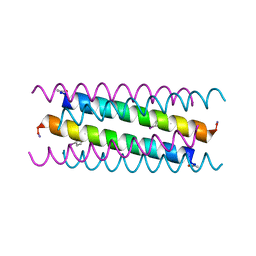 | |
5DPW
 
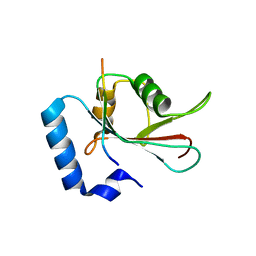 | |
5L1Q
 
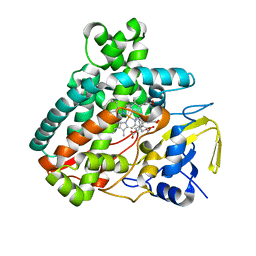 | | X-ray Structure of Cytochrome P450 PntM with Dihydropentalenolactone F | | Descriptor: | Dihydropentalenolactone F, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5HYM
 
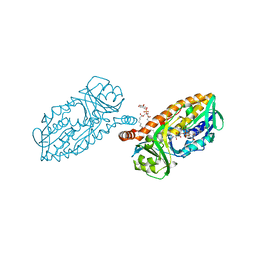 | | 3-Hydroxybenzoate 6-hydroxylase from Rhodococcus jostii in complex with phosphatidylinositol | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Phosphatidylinositol, ... | | Authors: | Orru, R, Montersino, S, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Hydroxybenzoate 6-Hydroxylase from Rhodococcus jostii RHA1 Contains a Phosphatidylinositol Cofactor.
Front Microbiol, 8, 2017
|
|
6XS3
 
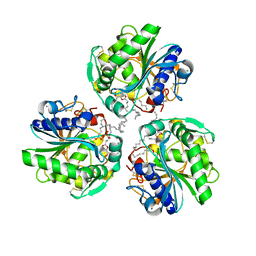 | |
5LBP
 
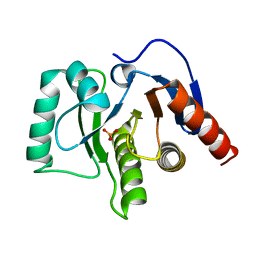 | | Oceanobacillus iheyensis macrodomain mutant N30A | | Descriptor: | MacroD-type macrodomain, PHOSPHATE ION | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
6Q3Z
 
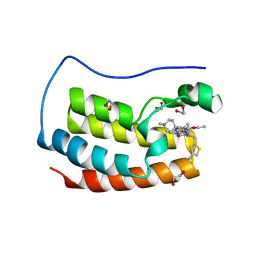 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16k | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-[(4-methylthiophen-2-yl)methyl]-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
6MYD
 
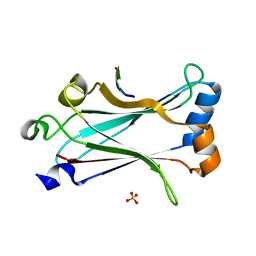 | | Structure of zebrafish TRAF6 in complex with STING CTT | | Descriptor: | STING CTT, Transmembrane protein 173, SULFATE ION, ... | | Authors: | de Oliveira Mann, C.C, Orzalli, M.H, King, D.S, Kagan, J.C, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2018-11-01 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Modular Architecture of the STING C-Terminal Tail Allows Interferon and NF-kappa B Signaling Adaptation.
Cell Rep, 27, 2019
|
|
6VFQ
 
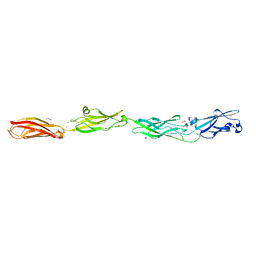 | | Crystal structure of monomeric human protocadherin 10 EC1-EC4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
5HQI
 
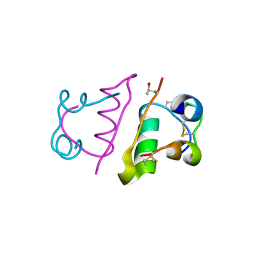 | | Insulin with proline analog HzP at position B28 in the T2 state | | Descriptor: | Insulin A-Chain, Insulin B-Chain | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
5E8W
 
 | |
8ONM
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with D-glutamate | | Descriptor: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing of the structural and catalytic roles of the residues in the active site of transaminase from Aminobacterium colombiense
To Be Published
|
|
6MZ1
 
 | |
6XT5
 
 | |
6Q43
 
 | | Atomic resolution crystal structure of an ABA collagen heterotrimer | | Descriptor: | GLYCEROL, Leading Chain of the ABA collagen heterotrimer, Middle Chain of the ABA collagen heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
