5TZG
 
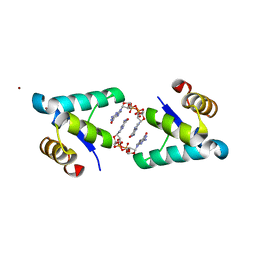 | | Structure of the BldD CTD(D116A)-(c-di-GMP)2, form 2 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein, ZINC ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
3O75
 
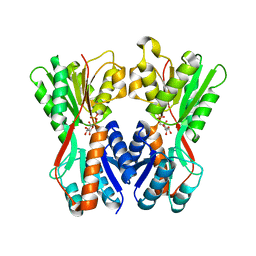 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida in complex with fructose 1-phosphate' | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, Fructose transport system repressor FruR | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
5TZD
 
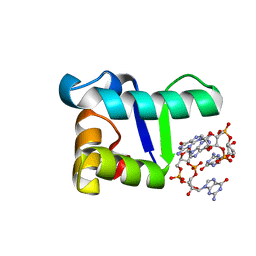 | | Structure of the WT S. venezulae BldD-(CTD-c-di-GMP)2 assembly intermediate | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
5H9T
 
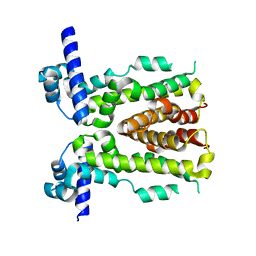 | | Crystal structure of native NalD at resolution of 2.9, the secondary repressor of MexAB-OprM multidrug efflux pump in Pseudomonas aeruginosa | | Descriptor: | NalD | | Authors: | Chen, W.Z, Wang, D, Huang, S.Q, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa.
Mol. Microbiol., 100, 2016
|
|
4YG4
 
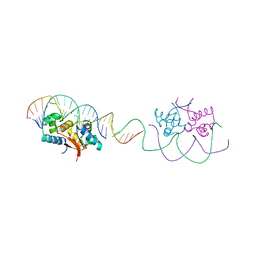 | | HipB-O1-O1* complex | | Descriptor: | Antitoxin HipB, DNA (28-MER), DNA (5'-D(*AP*TP*AP*TP*CP*CP*CP*CP*TP*TP*AP*AP*GP*GP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Schumacher, M.A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
1B1B
 
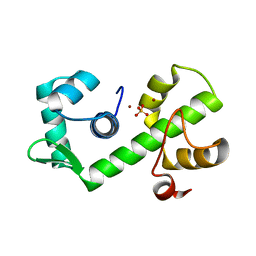 | | IRON DEPENDENT REGULATOR | | Descriptor: | PROTEIN (IRON DEPENDENT REGULATOR), SULFATE ION, ZINC ION | | Authors: | Pohl, E, Holmes, R.K, Hol, W.G. | | Deposit date: | 1998-11-19 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied.
J.Mol.Biol., 285, 1999
|
|
3PM1
 
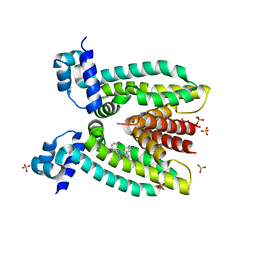 | | Structure of QacR E90Q bound to Ethidium | | Descriptor: | ETHIDIUM, HTH-type transcriptional regulator qacR, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A single acidic residue can guide binding site selection but does not govern QacR cationic-drug affinity.
Plos One, 6, 2011
|
|
4ZQD
 
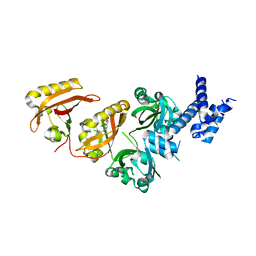 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex with the Benzoxadiazole Antagonist 0X3 | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(3-chloro-5-fluorophenyl)-4-nitro-2,1,3-benzoxadiazol-5-amine | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-09 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
2XB5
 
 | | Tet repressor (class D) in complex with 7-Iodotetracycline | | Descriptor: | 7-IODOTETRACYCLINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kisker, C, Saenger, W, Hinrichs, W. | | Deposit date: | 2010-04-05 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance.
Science, 264, 1994
|
|
2XPW
 
 | | TetR(D) in complex with oxytetracycline and magnesium. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Dalm, D, Proft, J, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-08-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Tetr(D) in Complex with Oxytetracycline.
To be Published
|
|
1HSJ
 
 | | SARR MBP FUSION STRUCTURE | | Descriptor: | FUSION PROTEIN CONSISTING OF STAPHYLOCOCCUS ACCESSORY REGULATOR PROTEIN R AND MALTOSE BINDING PROTEIN, alpha-D-glucopyranose | | Authors: | Zhang, G. | | Deposit date: | 2000-12-26 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the SarR protein from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8XAU
 
 | | Cryo-EM structure of HerA | | Descriptor: | ATP-binding protein | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8SFO
 
 | |
8XAV
 
 | | Cryo-EM structure of an anti-phage defense complex | | Descriptor: | ATP-binding protein, DUF4297 | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8SFN
 
 | |
4O8A
 
 | |
7S4A
 
 | | MRG15 complex with PALB2 peptide | | Descriptor: | Mortality factor 4-like protein 1, Partner and localizer of BRCA2 | | Authors: | Korolev, S, Deveryshetty, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Structural Insight into the Mechanism of PALB2 Interaction with MRG15.
Genes (Basel), 12, 2021
|
|
7RZY
 
 | | CryoEM structure of Vibrio cholerae transposon Tn6677 AAA+ ATPase TnsC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Tn6677 Vibrio cholerae transposon TnsC (VchTnsC) | | Authors: | Hoffmann, F.T, Kim, M, Beh, L.Y, Wang, J, Vo, P.L.H, Gelsinger, D.R, Acree, C, Mohabir, J.T, Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2021-08-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Nature, 609, 2022
|
|
2G0B
 
 | | The structure of FeeM, an N-acyl amino acid synthase from uncultured soil microbes | | Descriptor: | FeeM, N-DODECANOYL-L-TYROSINE | | Authors: | Van Wagoner, R.M, Clardy, J. | | Deposit date: | 2006-02-11 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | FeeM, an N-Acyl Amino Acid Synthase from an Uncultured Soil Microbe: Structure, Mechanism, and Acyl Carrier Protein Binding.
Structure, 14, 2006
|
|
4O9D
 
 | | Structure of Dos1 propeller | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Rik1-associated factor 1 | | Authors: | Kuscu, C, Schalch, T, Joshua-Tor, L. | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRL4-like Clr4 complex in Schizosaccharomyces pombe depends on an exposed surface of Dos1 for heterochromatin silencing.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6XLW
 
 | | Crystal structure of U2AF65 bound to AdML splice site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(P*UP*UP*(UD)P*UP*U)-D(P*(BRU))-R(P*CP*C)-3'), GLYCEROL, ... | | Authors: | Maji, D, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2020-06-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Representative cancer-associated U2AF2 mutations alter RNA interactions and splicing.
J.Biol.Chem., 295, 2020
|
|
5GTC
 
 | | Crystal structure of complex between DMAP-SH conjugated with a Kaposi's sarcoma herpesvirus LANA peptide (5-15) and nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Kato, D, Suto, H, Kurumizaka, H, Kawashima, S.A, Yamatsugu, K, Kanai, M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Posttranslational Modifications: Chemical Catalyst-Driven Regioselective Histone Acylation of Native Chromatin.
J. Am. Chem. Soc., 139, 2017
|
|
3Q3D
 
 | | Crystal structure of BmrR bound to puromycin | | Descriptor: | BmrR promoter DNA, Multidrug-efflux transporter 1 regulator, PUROMYCIN | | Authors: | Bachas, S, Eginton, C, Gunio, G, Wade, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural contributions to multidrug recognition in the multidrug resistance (MDR) gene regulator, BmrR.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1C1A
 
 | |
7SII
 
 | | Human STING bound to both cGAMP and 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide (Compound 53) | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, Stimulator of interferon genes protein, cGAMP | | Authors: | Lu, D, Shang, G, Jie, L, Lu, Y, Bai, X.C, Zhang, X. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Activation of STING by targeting a pocket in the transmembrane domain.
Nature, 604, 2022
|
|
