6Y6F
 
 | | Crystal structure of STK17B (DRAK2) in complex with PKIS43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-methylsulfanylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
5E1N
 
 | | Selenomethionine Ca2+-Calmodulin from Paramecium tetraurelia qFit disorder model | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calmodulin | | Authors: | Lin, J, van den Bedem, H, Brunger, A.T, Wilson, M.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution experimental phase information reveals extensive disorder and bound 2-methyl-2,4-pentanediol in Ca(2+)-calmodulin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8OQN
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-53 | | Descriptor: | 1-benzyl-1H-pyrazole-4-carboxylic acid, 3-hydroxyacyl-CoA dehydrogenase, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6G9M
 
 | |
5L5L
 
 | | Plexin A4 full extracellular region, domains 1 to 8 modeled, data to 8 angstrom, spacegroup P2(1) | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (8.001 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5FNM
 
 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | (~{E})-3-(4-methoxyphenyl)but-2-enoic acid, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-11-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
6PO7
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(3-(Aminomethyl)-4-(cyclopropylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-(cyclopropylmethoxy)phenyl]-4-methylquinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-03 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
5FJY
 
 | | Crystal structure of mouse kinesin light chain 2 (residues 161-480) | | Descriptor: | KINESIN LIGHT CHAIN 2, UNKNOWN PEPTIDE | | Authors: | Pernigo, S, Yip, Y.Y, Sanger, A, Xu, M, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Light Chains of Kinesin-1 are Autoinhibited.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6MV4
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR IXa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vadivel, K, Schreuder, H.A, Liesum, A, Bajaj, S.P. | | Deposit date: | 2018-10-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Sodium-site in serine protease domain of human coagulation factor IXa: evidence from the crystal structure and molecular dynamics simulations study.
J. Thromb. Haemost., 17, 2019
|
|
5FNS
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3s)-{4-Chloro-3-[(N-methylmethanesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl) propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
6PL0
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris in the Pr state bound to BV chromophore | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Sirigu, S, Klinke, S, Goldbaum, F, Chavas, L, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2019-06-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
6MVE
 
 | | Reduced X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
8OQL
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-1 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Hexafluorophosphate anion, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6MVG
 
 | |
6YAW
 
 | | Crystal structure of human GSTA1-1 bound to the glutathione adduct of cinnamaldehyde | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-[(1~{R})-3-oxidanylidene-1-phenyl-propyl]sulfanyl-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, GLYCEROL, Glutathione S-transferase A1 | | Authors: | Schwartz, M, Neiers, F. | | Deposit date: | 2020-03-13 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions Between Odorants and Glutathione Transferases in the Human Olfactory Cleft.
Chem.Senses, 45, 2020
|
|
6Y6N
 
 | | Structure of mature activin A with small molecule 2 | | Descriptor: | (3~{R})-3,4-dimethyl-3-propyl-1~{H}-1,4-benzodiazepine-2,5-dione, Inhibin beta A chain, SULFATE ION | | Authors: | McLoughlin, J.D, Brear, P.B, Reinhardt, T, Spring, D.R, Hyvonen, M. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6POV
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(3-(2-Aminoethyl)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)phenyl]-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
5E3N
 
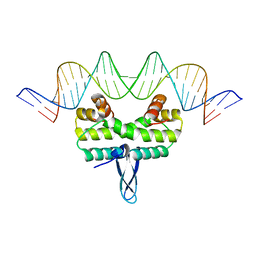 | |
6Y6Q
 
 | |
8OTL
 
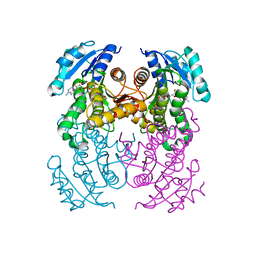 | | structure of InhA from Mycobacterium tuberculosis in complex with 5-(((4-(2-hydroxyphenoxy)benzyl)(octyl)amino)methyl)-2-phenoxyphenol | | Descriptor: | 1,2-ETHANEDIOL, 5-[[octyl-[[4-(2-oxidanylphenoxy)phenyl]methyl]amino]methyl]-2-phenoxy-phenol, ACETATE ION, ... | | Authors: | Tamhaev, R, Maveyraud, L, Chebaiki, M, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Exploring the plasticity of the InhA substrate-binding site using new diaryl ether inhibitors.
Bioorg.Chem., 143, 2023
|
|
8OI4
 
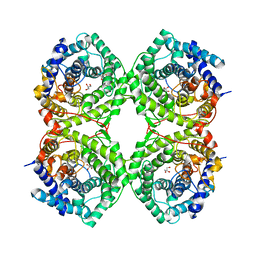 | | Metagenomic Beta-galactosidase from Glycoside Hydrolase family GH154 | | Descriptor: | Beta-galactosidase, CHLORIDE ION, GLYCEROL | | Authors: | Pijning, T, Hameleers, L, Jurak, E, Guskov, A. | | Deposit date: | 2023-03-22 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Novel beta-galactosidase activity and first crystal structure of Glycoside Hydrolase family 154.
N Biotechnol, 80, 2023
|
|
6V0Z
 
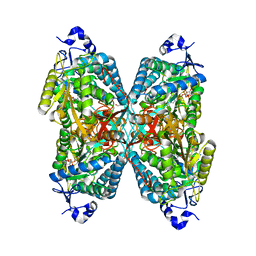 | | Structure of ALDH7A1 mutant R441C complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Korasick, D.A, Tanner, J.J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical, structural, and computational analyses of two new clinically identified missense mutations of ALDH7A1.
Chem.Biol.Interact., 2024
|
|
6MVT
 
 | | Structure of a bacterial ALDH16 complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Aldehyde dehydrogenase, SODIUM ION | | Authors: | Tanner, J.J, Liu, L. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J. Mol. Biol., 431, 2019
|
|
6FVZ
 
 | | Crystal structure of human monoamine oxidase B (MAO B) in complex with dimethylphenyl-chromone-carboxamide | | Descriptor: | Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Reis, J, Manzella, N, Cagide, F, Mialet-Perez, J, Uriarte, E, Parini, A, Borges, F, Binda, C. | | Deposit date: | 2018-03-05 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tight-Binding Inhibition of Human Monoamine Oxidase B by Chromone Analogs: A Kinetic, Crystallographic, and Biological Analysis.
J. Med. Chem., 61, 2018
|
|
8OSB
 
 | | TWIST1-TCF4-ALX4 complex on specific DNA | | Descriptor: | DNA (25-MER), Homeobox protein aristaless-like 4, Transcription factor 4, ... | | Authors: | Morgunova, E, Kim, S, Popov, A, Wysocka, J, Taipale, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-guided transcription factor cooperativity shapes face and limb mesenchyme.
Cell, 187, 2024
|
|
