7RU0
 
 | | SthK R120A Open State 1 | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, SthK | | Authors: | Gao, X, Nimigean, C, Schmidpeter, P. | | Deposit date: | 2021-08-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Gating intermediates reveal inhibitory role of the voltage sensor in a cyclic nucleotide-modulated ion channel.
Nat Commun, 13, 2022
|
|
7RYR
 
 | | SthK R120A Open State 3 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, SthK | | Authors: | Gao, X, Nimigean, C, Schmidpeter, P. | | Deposit date: | 2021-08-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Gating intermediates reveal inhibitory role of the voltage sensor in a cyclic nucleotide-modulated ion channel.
Nat Commun, 13, 2022
|
|
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4R8H
 
 | | The role of protein-ligand contacts in allosteric regulation of the Escherichia coli Catabolite Activator Protein | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, GLYCEROL, cAMP-activated global transcriptional regulator CRP | | Authors: | Townsend, P.D, Pohl, E, McLeish, T.C.B, Rodgers, T.L, Glover, L.C, Korhonen, H.J, Wilson, M.R, Hodgson, D.R.W, Cann, M.J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The Role of Protein-Ligand Contacts in Allosteric Regulation of the Escherichia coli Catabolite Activator Protein.
J.Biol.Chem., 290, 2015
|
|
7T8O
 
 | | Crystal Structure of the Crp/Fnr Family Transcriptional Regulator from Listeria monocytogenes | | Descriptor: | Lmo0753 protein, SULFATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Shatsman, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-16 | | Release date: | 2021-12-29 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of the Crp/Fnr Family Transcriptional Regulator from Listeria monocytogenes
To Be Published
|
|
4RZ7
 
 | | Crystal Structure of PVX_084705 with bound PCI32765 | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, UNKNOWN ATOM OR ION, cGMP-dependent protein kinase, ... | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal Structure of PVX_084705 with bound PCI32765
To be Published
|
|
1RUO
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP) MUTANT/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
1RUN
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Gunasekera, A, Vojtechovsky, J, Zhang, X, Kunkel, T.A, Berman, H.M, Ebright, R.H. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
6UQF
 
 | | Human HCN1 channel in a hyperpolarized conformation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MERCURY (II) ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
1I5Z
 
 | | STRUCTURE OF CRP-CAMP AT 1.9 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | White, M.A, Lee, J.C, Fox, R.O. | | Deposit date: | 2001-03-01 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The effect of the D53H point mutation on the macroscopic
motions of E. coli Cyclic AMP Receptor Protein
To be Published
|
|
6UQG
 
 | | Human HCN1 channel Y289D mutant | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
6V1X
 
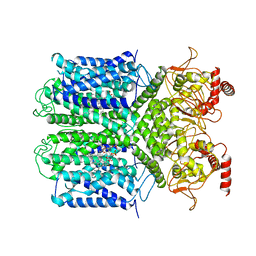 | | Cryo-EM Structure of the Hyperpolarization-Activated Potassium Channel KAT1: Tetramer | | Descriptor: | (2S)-1-(nonanoyloxy)-3-(phosphonooxy)propan-2-yl tetradecanoate, (3beta,5beta,14beta,17alpha)-cholestan-3-ol, Potassium channel KAT1 | | Authors: | Clark, M.D, Contreras, G.F, Shen, R, Perozo, E. | | Deposit date: | 2019-11-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Electromechanical coupling in the hyperpolarization-activated K + channel KAT1.
Nature, 583, 2020
|
|
4WBB
 
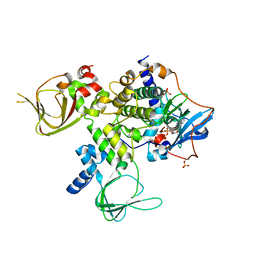 | | Single Turnover Autophosphorylation Cycle of the PKA RIIb Holoenzyme | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Zhang, P, Knape, M.J, Ahuja, L.G, Keshwani, M.M, King, C.C, Sastri, M, Herberg, F.W, Taylor, S.S. | | Deposit date: | 2014-09-02 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single Turnover Autophosphorylation Cycle of the PKA RII beta Holoenzyme.
Plos Biol., 13, 2015
|
|
6V1Y
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Potassium Channel KAT1: Octamer | | Descriptor: | (2S)-1-(nonanoyloxy)-3-(phosphonooxy)propan-2-yl tetradecanoate, (3beta,5beta,14beta,17alpha)-cholestan-3-ol, Potassium channel KAT1 | | Authors: | Clark, M.D, Contreras, G.F, Shen, R, Perozo, E. | | Deposit date: | 2019-11-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Electromechanical coupling in the hyperpolarization-activated K + channel KAT1.
Nature, 583, 2020
|
|
1I6X
 
 | | STRUCTURE OF A STAR MUTANT CRP-CAMP AT 2.2 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | White, M.A, Lee, J.C, Fox, R.O. | | Deposit date: | 2001-03-06 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The effect of the D53H point mutation on the macroscopic
motions of E. coli Cyclic AMP Receptor Protein
To be Published
|
|
1LB2
 
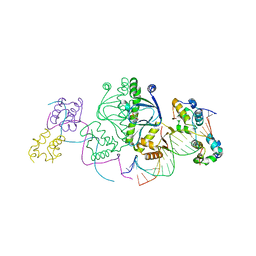 | | Structure of the E. coli alpha C-terminal domain of RNA polymerase in complex with CAP and DNA | | Descriptor: | 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*AP*G)-3', 5'-D(*CP*TP*TP*TP*TP*TP*TP*CP*CP*TP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Benoff, B, Yang, H, Lawson, C.L, Parkinson, G, Liu, J, Blatter, E, Ebright, Y.W, Berman, H.M, Ebright, R.H. | | Deposit date: | 2002-04-01 | | Release date: | 2002-09-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of transcription activation: the CAP-alpha CTD-DNA complex.
Science, 297, 2002
|
|
7ETS
 
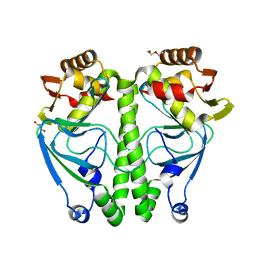 | | Crystal structure of crp protein from Gardnerella Vaginalis | | Descriptor: | Crp/Fnr family transcriptional regulator, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dong, H.J, Wang, S, Zhang, J.M, Gu, L. | | Deposit date: | 2021-05-13 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of crp protein from Gardnerella Vaginalis
To Be Published
|
|
7FEW
 
 | |
7FF0
 
 | |
7FF8
 
 | |
7FF9
 
 | |
6CJT
 
 | | Structure of the SthK cyclic nucleotide-gated potassium channel in complex with cGMP | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CYCLIC GUANOSINE MONOPHOSPHATE, SthK cyclic nucleotide-gated potassium channel | | Authors: | Nimigean, C.M, Rheinberger, J. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures.
Elife, 7, 2018
|
|
2HKX
 
 | | Structure of CooA mutant (N127L/S128L) from Carboxydothermus hydrogenoformans | | Descriptor: | CARBON MONOXIDE, Carbon monoxide oxidation system transcription regulator CooA-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lanz, N.D, Borjigin, M, Li, H, Kerby, R.L, Poulos, T.L, Roberts, G.P. | | Deposit date: | 2006-07-05 | | Release date: | 2007-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based hypothesis on the activation of the CO-sensing transcription factor CooA.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6CJQ
 
 | | Structure of the SthK cyclic nucleotide-gated potassium channel | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, SthK cyclic nucleotide-gated potassium channel | | Authors: | Nimigean, C.M, Rheinberger, J. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures.
Elife, 7, 2018
|
|
2GAU
 
 | | Crystal structure of transcriptional regulator, Crp/Fnr family from Porphyromonas gingivalis (APC80792), Structural genomics, MCSG | | Descriptor: | transcriptional regulator, Crp/Fnr family | | Authors: | Rotella, F.J, Zhang, R.G, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of transcriptional regulator, Crp/Fnr family from Porphyromonas gingivalis
To be Published
|
|
