1KQ3
 
 | | CRYSTAL STRUCTURE OF A GLYCEROL DEHYDROGENASE (TM0423) FROM THERMOTOGA MARITIMA AT 1.5 A RESOLUTION | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ZINC ION, ... | | Authors: | Wilson, I.A, Miller, M.D, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2002-01-03 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KQ4
 
 | |
1NF2
 
 | |
1K0S
 
 | | Solution structure of the chemotaxis protein CheW from the thermophilic organism Thermotoga maritima | | Descriptor: | CHEMOTAXIS PROTEIN CHEW | | Authors: | Griswold, I.J, Zhou, H, Swanson, R.V, Simon, M.I, Dahlquist, F.W. | | Deposit date: | 2001-09-20 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and interactions of CheW from Thermotoga maritima.
Nat.Struct.Biol., 9, 2002
|
|
1LME
 
 | | Crystal Structure of Peptide Deformylase from Thermotoga maritima | | Descriptor: | peptide deformylase | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2002-05-01 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1L2F
 
 | | Crystal structure of NusA from Thermotoga maritima: a structure-based role of the N-terminal domain | | Descriptor: | N utilization substance protein A | | Authors: | Shin, D.H, Nguyen, H.H, Jancarik, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-02-20 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain.
Biochemistry, 42, 2003
|
|
1KGS
 
 | |
1M4Y
 
 | | Crystal structure of HslV from Thermotoga maritima | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Song, H.K, Ramachandran, R, Bochtler, M.B, Hartmann, C, Azim, M.K, Huber, R. | | Deposit date: | 2002-07-05 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isolation and characterization of the prokaryotic proteasome homolog HslVU (ClpQY) from Thermotoga maritima and the crystal structure of HslV.
BIOPHYS.CHEM., 100, 2003
|
|
1MGP
 
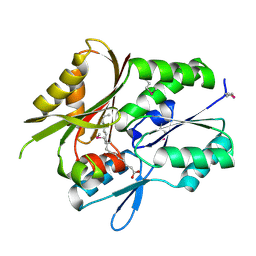 | | Hypothetical protein TM841 from Thermotoga maritima reveals fatty acid binding function | | Descriptor: | Hypothetical protein TM841, PALMITIC ACID | | Authors: | Schulze-Gahmen, U, Pelaschier, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-08-15 | | Release date: | 2002-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a hypothetical protein, TM841 of Thermotoga maritima,
reveals its function as fatty acid binding protein
Proteins, 50, 2003
|
|
3TMY
 
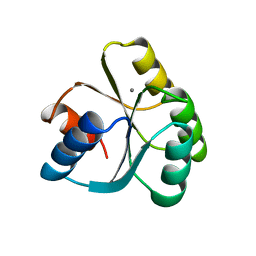 | | CHEY FROM THERMOTOGA MARITIMA (MN-III) | | Descriptor: | CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-04 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
1ZOR
 
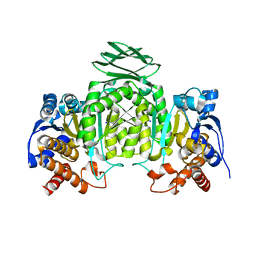 | | Isocitrate dehydrogenase from the hyperthermophile Thermotoga maritima | | Descriptor: | SODIUM ION, isocitrate dehydrogenase | | Authors: | Karlstrom, M, Steen, I.H, Birkeland, N.-K, Ladenstein, R. | | Deposit date: | 2005-05-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The crystal structure of a hyperthermostable subfamily II isocitrate dehydrogenase from Thermotoga maritima.
Febs J., 273, 2006
|
|
3S2C
 
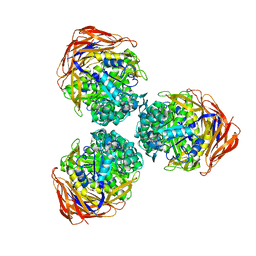 | | Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 | | Descriptor: | Alpha-N-arabinofuranosidase | | Authors: | Souza, T.A.C.B, Santos, C.R, Souza, A.R, Oldiges, D.P, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2011-05-16 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a novel thermostable GH51 Alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1
Protein Sci., 20, 2011
|
|
2B5D
 
 | | Crystal structure of the novel alpha-amylase AmyC from Thermotoga maritima | | Descriptor: | alpha-Amylase | | Authors: | Dickmanns, A, Ballschmiter, M, Liebl, W, Ficner, R. | | Deposit date: | 2005-09-28 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the novel alpha-amylase AmyC from Thermotoga maritima.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3UG4
 
 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3UG5
 
 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, beta-D-xylopyranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3UG3
 
 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, ... | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3T8Y
 
 | |
2BTY
 
 | | Acetylglutamate kinase from Thermotoga maritima complexed with its inhibitor arginine | | Descriptor: | ACETYLGLUTAMATE KINASE, ARGININE, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Gil-Ortiz, F, Fernandez-Murga, M.L, Fita, I, Rubio, V. | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Bases of Feed-Back Control of Arginine Biosynthesis, Revealed by the Structure of Two Hexameric N-Acetylglutamate Kinases, from Thermotoga Maritima and Pseudomonas Aeruginosa
J.Mol.Biol., 356, 2006
|
|
2K3F
 
 | |
2J73
 
 | |
1ODU
 
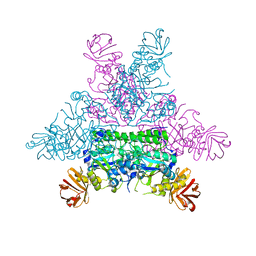 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA ALPHA-FUCOSIDASE IN COMPLEX WITH FUCOSE | | Descriptor: | PUTATIVE ALPHA-L-FUCOSIDASE, beta-L-fucopyranose | | Authors: | Sulzenbacher, G, Bignon, C, Bourne, Y, Henrissat, B. | | Deposit date: | 2003-03-14 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Thermotoga Maritima {Alpha}-L-Fucosidase: Insights Into the Catalytic Mechanism and the Molecular Basis for Fucosidosis
J.Biol.Chem., 279, 2004
|
|
1Z77
 
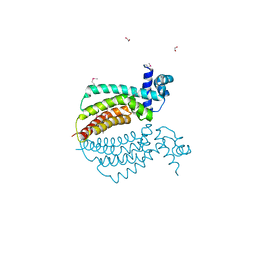 | | Crystal structure of transcriptional regulator protein from Thermotoga maritima. | | Descriptor: | 1,2-ETHANEDIOL, transcriptional regulator (TetR family) | | Authors: | Koclega, K.D, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Kudritska, M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-24 | | Release date: | 2005-05-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a transcriptional regulator TM1030 from Thermotoga maritima solved by an unusual MAD experiment.
J.Struct.Biol., 159, 2007
|
|
3U95
 
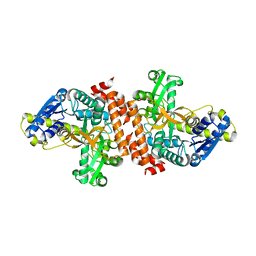 | | Crystal structure of a putative alpha-glucosidase from Thermotoga neapolitana | | Descriptor: | Glycoside hydrolase, family 4, MANGANESE (II) ION | | Authors: | Ha, N.C, Jun, S.Y, Yun, B.Y, Yoon, B.Y, Piao, S. | | Deposit date: | 2011-10-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure and thermostability of a putative alpha-glucosidase from Thermotoga neapolitana
Biochem.Biophys.Res.Commun., 416, 2011
|
|
2A6A
 
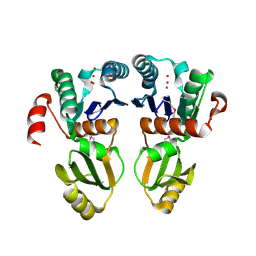 | |
2GAI
 
 | |
