2A9I
 
 | |
1VWI
 
 | |
1VWN
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 4.8 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWJ
 
 | |
1VWE
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 3.6 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
4S21
 
 | | Crystal structure of the photosensory core module of bacteriophytochrome RPA3015 from R. palustris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome (Light-regulated signal transduction histidine kinase), PhyB1 | | Authors: | Yang, X, Stojkovi, E.A, Ozarowski, W.B, Moffat, K. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Light Signaling Mechanism of Two Tandem Bacteriophytochromes.
Structure, 23, 2015
|
|
4I6W
 
 | | 3-hydroxy-3-methylglutaryl (HMG) Coenzyme-A reductase complexed with thiomevalonate | | Descriptor: | (3S)-3-hydroxy-3-methyl-5-sulfanylpentanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, ... | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Burgner II, J.W, Rodwell, V.W, Wrensford, L.V, Critchelow, C.J, Min, J. | | Deposit date: | 2012-11-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
4I3A
 
 | | Structures of PR1 and PR2 intermediates from time-resolved laue crystallography collected at 14ID-B, APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4TX3
 
 | | Complex of the X-domain and OxyB from Teicoplanin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, OxyB protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Peschke, M, Haslinger, K, Cryle, M.J. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis.
Nature, 521, 2015
|
|
4JK1
 
 | |
2A1A
 
 | | PKR kinase domain-eIF2alpha Complex | | Descriptor: | Eukaryotic translation initiation factor 2 alpha subunit, Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Dar, A.C, Dever, T.E, Sicheri, F. | | Deposit date: | 2005-06-19 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Higher-Order Substrate Recognition of eIF2alpha by the RNA-Dependent Protein Kinase PKR.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A2S
 
 | | Crystal Structure of Human Glutathione Transferase in complex with S-nitrosoglutathione in the absence of reducing agent | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5-[1-(CARBOXYLATOMETHYLCARBAMOYL)-2-NITROSOSULFANYL-ETHYL]AMINO-5-OXO-PENTANOATE, CALCIUM ION, ... | | Authors: | Parker, L.J, Morton, C.J, Adams, J.J, Parker, M.W. | | Deposit date: | 2005-06-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1
Protein Sci., 15, 2006
|
|
4JT0
 
 | | Yeast 20S proteasome in complex with the dimerized linear mimetic of TMC-95A - yCP:4a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Desvergne, A, Genin, E, Marechal, X, Gallastegui, N, Dufau, L, Richy, N, Groll, M, Vidal, J, Reboud-Ravaux, M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome.
J.Med.Chem., 56, 2013
|
|
2A7P
 
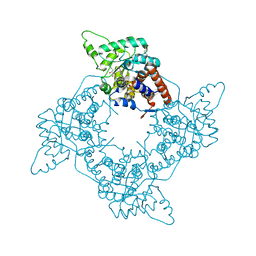 | | Crystal Structure of the G81A mutant of the Active Chimera of (S)-Mandelate Dehydrogenase in complex with its substrate 3-indolelactate | | Descriptor: | (S)-Mandelate Dehydrogenase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(INDOL-3-YL) LACTATE, ... | | Authors: | Sukumar, N, Xu, Y, Mitra, B, Mathews, F.S. | | Deposit date: | 2005-07-05 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the G81A mutant form of the active chimera of (S)-mandelate dehydrogenase and its complex with two of its substrates.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2A8K
 
 | |
2AAS
 
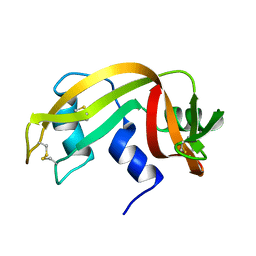 | | HIGH-RESOLUTION THREE-DIMENSIONAL STRUCTURE OF RIBONUCLEASE A IN SOLUTION BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | RIBONUCLEASE A | | Authors: | Santoro, J, Gonzalez, C, Bruix, M, Neira, J.L, Nieto, J.L, Herranz, J, Rico, M. | | Deposit date: | 1992-11-20 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution three-dimensional structure of ribonuclease A in solution by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 229, 1993
|
|
2AB4
 
 | |
2AEW
 
 | |
2A0Q
 
 | | Structure of thrombin in 400 mM potassium chloride | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Papaconstantinou, M, Carrell, C.J, Pineda, A.O, Bobofchak, K.M, Mathews, F.S, Flordellis, C.S, Maragoudakis, M.E, Tsopanoglou, N.E, di Cera, E. | | Deposit date: | 2005-06-16 | | Release date: | 2005-07-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thrombin Functions through Its RGD Sequence in a Non-canonical Conformation.
J.Biol.Chem., 280, 2005
|
|
2A3F
 
 | | Crystal structure of nitrophorin 2 aqua complex | | Descriptor: | Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus.
To be Published
|
|
2A6H
 
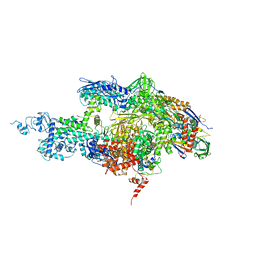 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic sterptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Temiakov, D, Zenkin, N, Vassylyeva, M.N, Perederina, A, Tahirov, T.H, Savkina, M, Zorov, S, Nikiforov, V, Igarashi, N, Matsugaki, N, Wakatsuki, S, Severinov, K, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of transcription inhibition by antibiotic streptolydigin.
Mol.Cell, 19, 2005
|
|
1Z7E
 
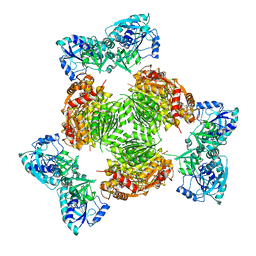 | | Crystal structure of full length ArnA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID, protein ArnA | | Authors: | Gatzeva-Topalova, P.Z, May, A.P, Sousa, M.C. | | Deposit date: | 2005-03-24 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and Mechanism of ArnA: Conformational Change Implies Ordered Dehydrogenase Mechanism in Key Enzyme for Polymyxin Resistance
Structure, 13, 2005
|
|
4IRB
 
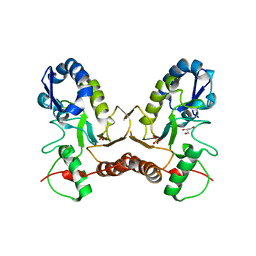 | | Crystal Structure of Vaccinia Virus Uracil DNA Glycosylase Mutant del171-172D4 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Schormann, N, Zhukovskaya, N, Sartmatova, D, Nuth, M, Ricciardi, R.P, Chattopadhyay, D. | | Deposit date: | 2013-01-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at the dimer interface affect both function and structure of the Vaccinia virus uracil DNA glycosylase
To be Published
|
|
1ZM3
 
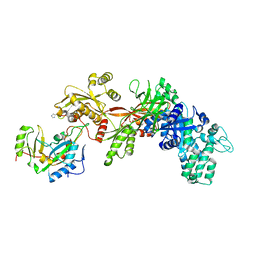 | | Structure of the apo eEF2-ETA complex | | Descriptor: | Elongation factor 2, exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
