3H16
 
 | | Crystal structure of a bacteria TIR domain, PdTIR from Paracoccus denitrificans | | Descriptor: | SULFATE ION, TIR protein | | Authors: | Chan, S.L, Low, L.Y, Santelli, E, Pascual, J. | | Deposit date: | 2009-04-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mimicry in Innate Immunity: CRYSTAL STRUCTURE OF A BACTERIAL TIR DOMAIN.
J.Biol.Chem., 284, 2009
|
|
3R52
 
 | | Structure analysis of a wound-inducible lectin ipomoelin from sweet potato | | Descriptor: | CADMIUM ION, Ipomoelin, methyl alpha-D-glucopyranoside | | Authors: | Liu, K.L, Chang, W.C, Jeng, S.T, Cheng, Y.S. | | Deposit date: | 2011-03-18 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ipomoelin, a jacalin-related lectin with a compact tetrameric association and versatile carbohydrate binding properties regulated by its N terminus.
Plos One, 7, 2012
|
|
2VJU
 
 | | Crystal structure of the IS608 transposase in complex with the complete Right end 35-mer DNA and manganese | | Descriptor: | MANGANESE (II) ION, RIGHT END 35-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VCP
 
 | | Crystal structure of N-Wasp VC domain in complex with skeletal actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gaucher, J.F, Didry, D, Carlier, M.F. | | Deposit date: | 2007-09-26 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Interactions of isolated C-terminal fragments of neural Wiskott-Aldrich syndrome protein (N-WASP) with actin and Arp2/3 complex.
J. Biol. Chem., 287, 2012
|
|
2VGE
 
 | | Crystal structure of the C-terminal region of human iASPP | | Descriptor: | RELA-ASSOCIATED INHIBITOR | | Authors: | Robinson, R.A, Lu, X, Jones, E.Y, Siebold, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Studies of Aspp Proteins Reveal Differential Binding to P53, P63 and P73
Structure, 16, 2008
|
|
3H73
 
 | |
2VH2
 
 | | Crystal structure of cell divison protein FtsQ from Yersinia enterecolitica | | Descriptor: | CELL DIVISION PROTEIN FTSQ | | Authors: | van den Ent, F, Vinkenvleugel, T, Ind, A, West, P, Veprintsev, D, Nanninga, N, den Blaauwen, T, Lowe, J. | | Deposit date: | 2007-11-16 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and Mutational Analysis of Cell Division Protein Ftsq
Mol.Microbiol., 68, 2008
|
|
2VQ0
 
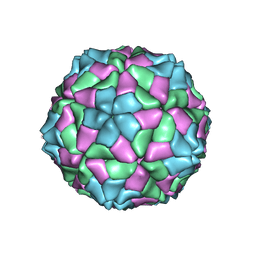 | | Capsid structure of Sesbania mosaic virus coat protein deletion mutant rCP(delta 48 to 59) | | Descriptor: | CALCIUM ION, COAT PROTEIN | | Authors: | Anju, P, Subashchandrabose, C, Satheshkumar, P.S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of Recombinant Capsids Formed by the Beta-Annulus Deletion Mutant-Rcp (Delta 48-59) of Sesbania Mosaic Virus
Virology, 375, 2008
|
|
8PIU
 
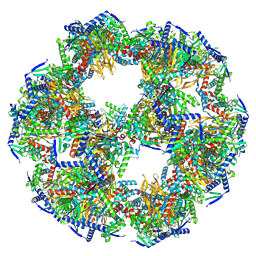 | |
3E1R
 
 | | Midbody targeting of the ESCRT machinery by a non-canonical coiled-coil in CEP55 | | Descriptor: | Centrosomal protein of 55 kDa, Programmed cell death 6-interacting protein | | Authors: | Lee, H.H, Elia, N, Ghirlando, R, Lippincott-Schwartz, J, Hurley, J.H. | | Deposit date: | 2008-08-04 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55.
Science, 322, 2008
|
|
2VPX
 
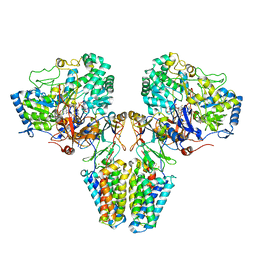 | | Polysulfide reductase with bound quinone (UQ1) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3DBV
 
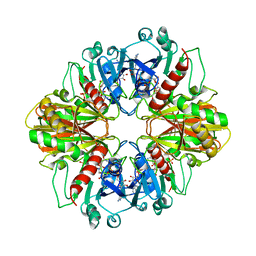 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
2VT7
 
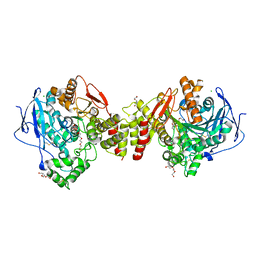 | | Native Torpedo californica acetylcholinesterase collected with a cumulated dose of 800000 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Colletier, J.P, Bourgeois, D, Sanson, B, Fournier, D, Sussman, J.L, Silman, I, Weik, M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3DAA
 
 | |
3ER5
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, H-189 | | Authors: | Bailey, D, Veerapandian, B, Cooper, J, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray-crystallographic studies of complexes of pepstatin A and a statine-containing human renin inhibitor with endothiapepsin.
Biochem.J., 289 ( Pt 2), 1993
|
|
3E23
 
 | | Crystal structure of the RPA2492 protein in complex with SAM from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR299 | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, uncharacterized protein RPA2492 | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Mao, L, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, H, Tong, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-05 | | Release date: | 2008-09-30 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the RPA2492 protein in complex with SAM from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR299
To be Published
|
|
2W06
 
 | | Structure of CDK2 in complex with an imidazolyl pyrimidine, compound 5c | | Descriptor: | 4-{[4-(1-CYCLOPROPYL-2-METHYL-1H-IMIDAZOL-5-YL)PYRIMIDIN-2-YL]AMINO}-N-METHYLBENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Anderson, M, Andrews, D.M, Barker, A.J, Brassington, C.A, Byth, K.F, Culshaw, J.D, Finlay, M.R.V, Fisher, E, Mcmiken, H.H.J, Green, C.P, Heaton, D.W, Nash, I.A, Newcombe, N.J, Oakes, S.E, Roberts, A, Stanway, J.J, Thomas, A.P, Tucker, J.A, Weir, H.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Imidazoles: Sar and Development of a Potent Class of Cyclin-Dependent Kinase Inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VOP
 
 | | Crystal structure of N-terminal domains of Human La protein complexed with RNA oligomer AUUUU | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP)-3', LUPUS LA PROTEIN, SULFATE ION | | Authors: | Kotik-Kogan, O, Valentine, E.R, Sanfelice, D, Conte, M.R, Curry, S. | | Deposit date: | 2008-02-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis Reveals Conformational Plasticity in the Recognition of RNA 3' Ends by the Human La Protein.
Structure, 16, 2008
|
|
8PJL
 
 | |
2VZ3
 
 | | bleached galactose oxidase | | Descriptor: | ACETATE ION, COPPER (II) ION, GALACTOSE OXIDASE | | Authors: | Rogers, M.S, Hurtado-Guerrero, R, Firbank, S.J, Halcrow, M.A, Dooley, D.M, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Link Formation of the Cysteine 228-Tyrosine 272 Catalytic Cofactor of Galactose Oxidase Does not Require Dioxygen.
Biochemistry, 47, 2008
|
|
2VQK
 
 | |
3EEG
 
 | |
2VV1
 
 | | hPPARgamma Ligand binding domain in complex with 4-HDHA | | Descriptor: | (4S,5E,7Z,10Z,13Z,16Z,19Z)-4-hydroxydocosa-5,7,10,13,16,19-hexaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VS5
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GALACTOSE-URIDINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
3U9S
 
 | | Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Methylcrotonyl-CoA carboxylase, ... | | Authors: | Huang, C.S, Tong, L. | | Deposit date: | 2011-10-19 | | Release date: | 2011-12-14 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An unanticipated architecture of the 750-kDa {alpha}6{beta}6 holoenzyme of 3-methylcrotonyl-CoA carboxylase
Nature, 481, 2012
|
|
